Size exclusion chromatography (SEC) is a proven method for molecular weight (MW or molar mass) measurement of proteins, typically involving comparison of the column retention volume of a sample against a string of known MW standards.
Since the hydrodynamic volume of a molecule (not the molecular weight) actually controls the separation process in SEC, the ensuing results are only relative to the standards employed in the calibration of the system.
How close the "relative" MW is to the actual MW of a sample is influenced by the extent of similarity between the standards and analyte structures. Hence, any difference in the structure will have a negative impact on the accuracy of the results.
Additionally, the interaction of some proteins with the column matrix and their tendency to alter hydrodynamic volume during elution make the retention volume ineffective in the calculation of the MW of the measured molecule.
The integration of sophisticated detectors with chromatography systems enables measuring the absolute MW of a sample independent of any standards or sample elution volume.
This article presents two examples demonstrating the inapplicability of the retention volume of a sample to obtain accurate molecular weight measurements. It also describes the ability of the advanced detectors to obtain additional information.
Instrumentation
A Viscotek TDAmax system, comprising a Triple Detector Array (TDA) connected to a GPCmax degasser, autosampler module, pump and SEC column, was used in these examples. Bovine serum albumin (BSA) injections were employed in the determination of inter-detector delay volumes, band broadening correction factors, and calibration constants. The OmniSEC software was employed for controlling the whole system and acquiring the signals from the five detectors of the TDA system.
The five detectors of the TDA system include a differential pressure (DP) Wheatstone bridge viscometer, a differential refractometer (RI), 90° Right Angle Light Scattering (RALS) and 7° Low Angle Light Scattering (LALS) detectors quantifying the static light scattering intensity, and a UV-Visible spectrophotometer.
The absolute molecular weight (MW) and the intrinsic viscosity (IV) of the protein were calculated from the measurements by the five detectors using the OmniSEC software.
Experimental Results
Absolute Molecular Weight Measurement Following Sample-Matrix Interaction
In this analysis, Anthrolysin (ALO) was studied. The SEC experiment used a Superdex 200 (GE Healthcare) with a buffer consisting of 150 mM NaCl, 20 mM Tris.
The MW and IV of ALO in solution were determined using the Viscotek TDA and the results are presented in Figures 1A and 1B. Data lines in Figure 1A show UV (purple), RALS (green), LALS (black), and DP (blue). B -molecular weight (MW) and intrinsic viscosity (IV) patterns of ALO. Data lines in Figure 1B show molecular weight (black), UV (purple), and IV (grey).
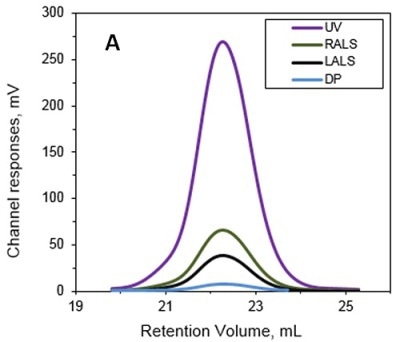
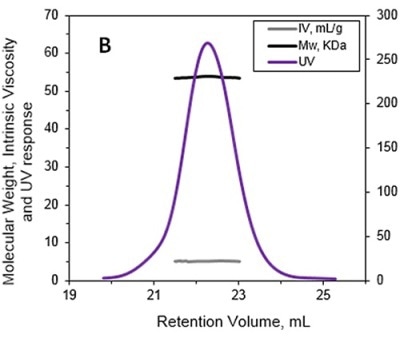
Figure 1. A. Chromatograms of ALO from the Viscotek TDAmax. B. Molecular weight (MW) and intrinsic viscosity (IV) patterns of ALO.
The calculated absolute MW was 53.6 kDa, revealing that ALO exists in monomeric form when in solution. Conversely, the MW measurement with a conventional column calibration method would yield a value of only 15-20 kDa, thus necessitating the use of light scattering detectors to avoid such underestimation of MW.
The IV of the sample was determined using the online differential viscometer. The measured IV value for ALO was 5.1 mL/g. The comparison with a BSA monomer having a MW of 66.5 kDa and IV of roughly 3.2 mL/g reveals that ALO has a lower density and more open structure.
IV measurements can be utilized in the calculation of the hydrodynamic radius of a sample. The fact that ALO is higher than would be expected than BSA reveals the interaction with the column.
Influence of Sample Structure on Sample Elution Volume
Conventional SEC analyzes are incapable of justifying the structural differences between the analyte of interest and the standard. This example was the in vitro formation of aggregates by a Bcl-2 protein by incubating the protein at 37°C. The early events happening during the aggregation process subsequent to incubation for 1 day and 1 week were characterized using SEC with a Superose 6HR (GE Healthcare), 150 mM sodium chloride, and a buffer of 20 mM sodium phosphate.
A characteristic triple detection chromatogram after one day of incubation at 37°C is depicted in Figure 2. Inset reveals the MW distribution of the different components within the sample.
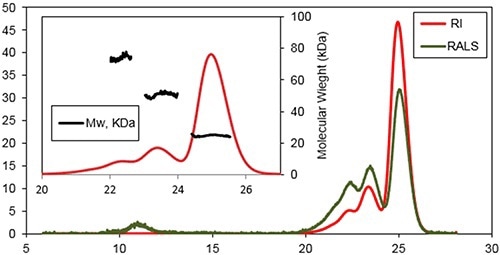
Figure 2. Chromatogram of Bcl-2 from the Viscotek TDAmax. RALS (green line) and Refractive Index detector (red line).
Three main species were determined at the retention volumes of 22.6, 23.6, and 25.1 mL with MWs of 74±3 kDa, 51±2 kDa, and 25±2 kDa, respectively. The monomeric form of the protein was the main population of this sample.
A shoulder was observable on the trimers peak, representing a small population of tetramer. Additional species were determined when eluting at 11 mL by the light scattering detectors. This is a characteristic response for protein aggregates with a very high MW, but with a low concentration.
The MW measurement of the Bcl-2 monomer with a conventional column calibration method would yield a value of only 40 kDa, suggesting that the Bcl-2 has higher hydrodynamic radius (RH) than expected although it is monomeric. Assuming that the sample has no interaction with column matrix, this effect could take place because of a deviation from the spherical shape and/or hydration of the protein.
The analysis of the sample incubated for one week at 37°C revealed the formation of more aggregates. According to the SEC analyzes, the formation of small aggregates (mostly dimers and trimers) was the first step of the fiber growth. These small aggregates further assemble into larger aggregates.
Conclusions
SEC is a useful technique to gain insights into proteins and their functionality. This article discussed the shortcomings of using conventional column calibration techniques in molecular weight measurements.
The results demonstrate the ability of a multi-detection system to determine the absolute MW of a protein and its oligomers irrespective of the retention volumes or any standards utilized for calibration.
They also show that this sophisticated method is capable of differentiating between extent of aggregation, oligomeric state, shape differences or hydration within the sample.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.