It is essential for manufacturers to have an understanding about the effects of processing a polymer on its molecular weight distribution, molecular weight and solution viscosity. Time and money can be saved by learning whether a reground or recycled polymer will still fall within specifications. Knowing what the polymer characteristics are at end use can help prevent failures and enhance specifications.
This article describes the Gel Permeation Chromatography (GPC) or equivalently Size Exclusion Chromatography (SEC), analysis of the degradation of poly(lactic acid) (PLA) in both of these described scenarios. However, the general analysis and methods can be applied to any polymer system where degradation could be an problem.
Experiment
Two different sets of PLA samples were examined in this article. The first set focuses on the effect of regrinding the polymer after an initial use for recycling. With this analysis, polymer manufacturers will be able to know if and how many times they can recycle their product while remaining within the set specifications. The polymers will be referred to as Regrind and Virgin.
The second set focuses on the effect of 3D printing on a PLA feedstock and the final printed product. This analysis aims to explain the stability of the feedstock to the printing process. With this information, a user will be able to adjust the incoming feedstock to either account for the expected degradation or minimize degradation (with additives or stabilizers). These polymers will be referred to as 1-Pre, 1-Post, 2-Pre, and 2-Post, where “1” and “2” refer to the varied PLA formulations and “Pre” and “Post” refer to the polymers before and after printing.
A Malvern Panalytical OMNISEC RESOLVE and REVEAL triple detection GPC system was used to examine the two different sets of polymers, as shown in Figure 1. The system is provided with a light scattering detector to directly measure the molecular weight (MW), a viscometer detector to measure the intrinsic viscosity (IV) of the sample and a refractive index (RI) detector to measure the concentration at each elution time.

Figure 1. Malvern Panalytical’s OMNISEC Triple Detection GPC/SEC System.
Effect of Regrinding on Molecular Characteristics
Figures 2 and 3 depict the triple detector chromatograms for samples Virgin and Regrind, respectively. In these chromatograms, the RI detector signal is shown in red, the right angle light scattering (RALS) signal is shown in green and the viscometer signal is shown in blue. The calculated MW at each slice of the chromatogram is indicated with the gold curve.
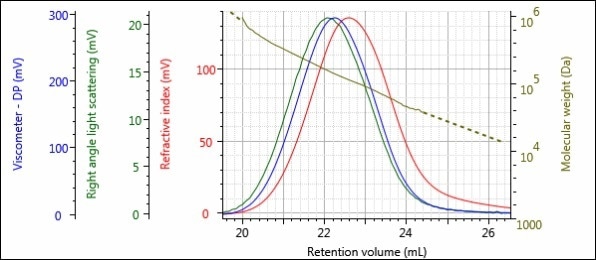
Figure 2. Triple detector chromatogram of the Virgin sample.
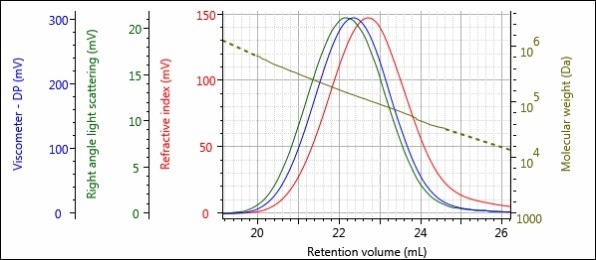
Figure 3. Triple detector chromatogram of the Regrind sample.
At first glance, these two injections seem to be almost identical. A helpful way to better compare these two samples would be to overlay them on the same plot as illustrated in Figure 4.
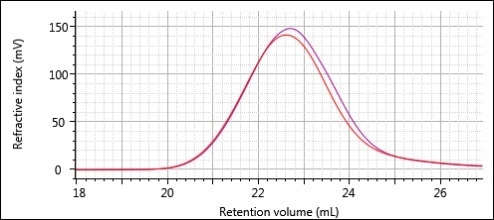
Figure 4. Overlay of the RI chromatograms for the Virgin (red) and Regrind (purple) samples.
There is a slight difference from inspection of Figure 4. The Regrind sample seems to have an increased amount of material on the tailing end of the peak, which corresponds to lower MW polymer. By taking the quantitative results from the software and comparing those, it is possible to exactly determine how much difference there is between the two samples. These results are summarized in Table 1 below. The data shown is an average of three injections of each sample. Presented values include the moments of the MW distribution (Mn, Mw, and Mz), hydrodynamic radius (Rh) and dispersity (Mw/Mn), IV.
Table 1. Calculated results for PLA samples before and after regrinding
| |
VIRGIN |
REGRIND |
| Average |
St Dev |
Average |
St Dev |
| Mn (g/mol) |
84,283 |
2,180 |
72,797 |
1,507 |
| Mw (g/mol) |
130,167 |
1,301 |
124,800 |
529 |
| Mz (g/mol) |
190,033 |
1,617 |
180,533 |
1,498 |
| Dispersity |
1.545 |
0.025 |
1.715 |
0.029 |
| IV (dL/g) |
1.547 |
0.030 |
1.510 |
0.004 |
| Rh (nm) |
14.18 |
0.15 |
13.88 |
0.04 |
This data highlights that the samples are degrading in that the MW is dropping along with the IV and Rh. While the differences are small, they are still statistically different and this could be the difference between a product being within a given specification or not.
Effect of 3D Printing on Molecular Characteristics
Figures 5 – 8 show the triple detector chromatograms of samples 1-Pre, 1-Post, 2-Pre and 2-Post. As with Figures 2 and 3, the RI detector signal is shown in red, the RALS signal is shown in green, the viscometer signal is shown in blue and the calculated MW at each slice of the chromatogram is shown with the gold curve.
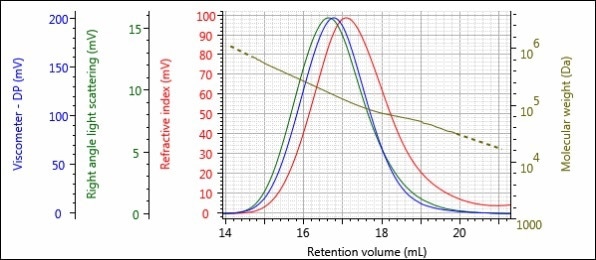
Figure 5. Triple detector chromatogram of sample 1-Pre.
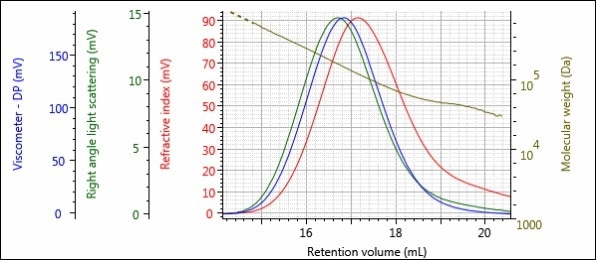
Figure 6. Triple detector chromatogram of sample 1-Post.
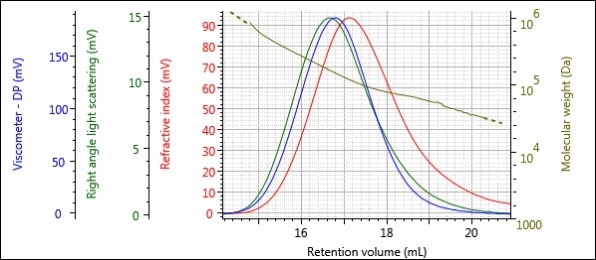
Figure 7. Triple detector chromatogram of sample 2-Pre.
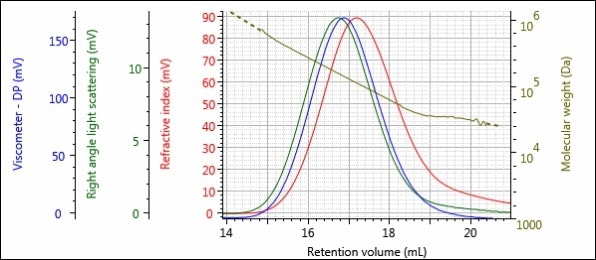
Figure 8. Triple detector chromatogram of sample 2-Post.
The overlays of the RI chromatograms for the two different formulations are shown in Figures 9 and 10. For both figures, the polymers before printing and after printing are in red and purple, respectively.
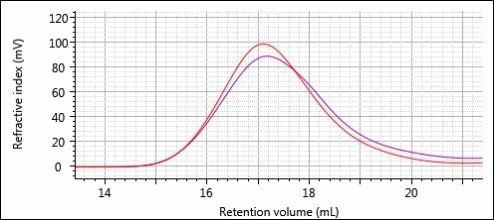
Figure 9. Overlay of RI chromatograms for samples 1-Pre (red) and 1-Post (purple).
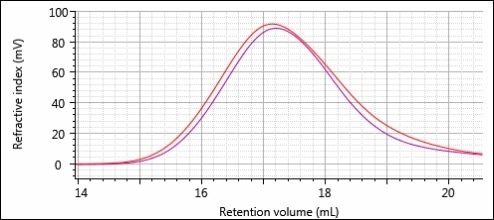
Figure 10. Overlay of the RI chromatograms for samples 2-Pre (red) and 2-Post (purple).
Distinct differences can be seen between the peak shapes for formulation 1 (Figure 9). There is more material on the back end of the peak after printing and more polymer on the front end of the peak before printing. From this, one can infer a decrease in MW due to printing. There are more differences in peak intensity rather than peak shape, while there are differences between the two chromatograms for formulation 2 (Figure 10). This peak intensity difference is most likely from a difference in concentration between the two sample solutions prepared for analysis. As before, it is possible to perform a more detailed and accurate comparison using the quantitative results shown in Table 2.
Table 2. Calculated results for the two PLA formulations before and after 3D printing
| |
Formulation 1 |
Formulation 2 |
| 1-Pre |
1-Post |
2-Pre |
2-Post |
| Average |
St Dev |
Average |
St Dev |
Average |
St Dev |
Average |
St Dev |
| Mn (g/mol) |
93,272 |
2,345 |
89,313 |
2,209 |
95,497 |
1,108 |
82,133 |
655 |
| Mw (g/mol) |
135,967 |
907 |
130,400 |
200 |
133,500 |
1,015 |
122,800 |
600 |
| Mz (g/mol) |
197,900 |
2,052 |
185,233 |
2,857 |
191,033 |
3,202 |
180,933 |
907 |
| Dispersity |
1.458 |
0.028 |
1.461 |
0.035 |
1.398 |
0.014 |
1.496 |
0.007 |
| IV (dL/g) |
1.218 |
0.010 |
1.155 |
0.026 |
1.109 |
0.044 |
1.107 |
0.040 |
| Rh (nm) |
13.26 |
0.08 |
12.79 |
0.10 |
12.67 |
0.24 |
12.32 |
0.21 |
Both formulations show a decrease in IV, MW and Rh after printing. Formulation 2 shows a more pronounced decrease in MW both relatively and in absolute magnitude, despite peak shapes being fairly similar. This type of information can be valuable to manufacturers looking for a polymer to better withstand the printing process.
Conclusion
GPC/SEC provides an efficient method to determine polymer degradation. A user can determine changes in MW, IV and Rh by analyzing the sample before and after processing. These metrics and changes in these metrics are very significant to manufacturers seeking to recycle some unused polymer into the feedstock or looking into issues with a final, processed product. This increases profit margins by reducing wasted product and limiting the requirement for recalls and/or manufacturing downtime to deal with issues.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.