Jul 18 2016
University of Basel chemists have succeeded in applying computer simulations to explain transient structures in proteins. The journal Angewandte Chemie features a report in which the chemists illustrate the possibility of understanding proteins’ modes of action by using computer simulations of information at the atomic level.
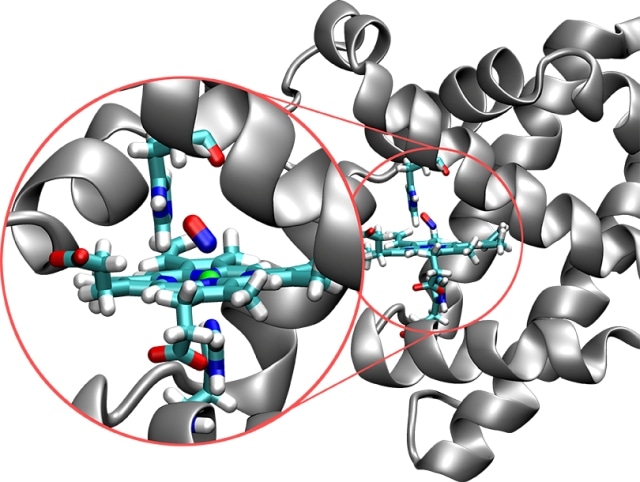 Structure of the protein myoglobin (silver) with the embedded active site (in color). In this image, the nitrogen molecule (red/blue) is bonded to the iron atom (green ball). (CREDIT: University of Basel)
Structure of the protein myoglobin (silver) with the embedded active site (in color). In this image, the nitrogen molecule (red/blue) is bonded to the iron atom (green ball). (CREDIT: University of Basel)
The movement of individual atoms of a molecule can be characterized by using computational chemistry. The simulation techniques that are currently used enable scientists to explain the dynamics of molecules and systems with hundreds of thousands of atoms. These simulation techniques also play a significant role in characterizing molecular states that cannot be easily and directly observed in experiments because of their short lifetime. In this situation, computer simulations are considered to be a valuable source providing complementary insight.
The dynamics and structure of a protein helps determining its function. It is important to obtain details related to the molecular processes and the nature of structures in the active site, which refers to the place where chemical reactions occur. Structural changes are caused during the development and breaking of chemical bonds. Generally, the observable dynamics result in steady (low energy) states that are attained through one or more metastable (higher energy) intermediate procedures. The possibility of directly identifying a metastable state in an experiment relies on its lifetime. A short lifespan results in only indirect detection methods.
Computer determines atomic geometry
Recently, a team of researchers headed by Prof. Markus Meuwly from the Department of Chemistry at the University of Basel characterized the temporal and spatial behavior of the protein myoglobin by using molecular dynamics simulations. Myoglobin plays a vital role in transferring oxygen within cells and is mostly present in muscle tissue. Nitrogen monoxide is a reactive and short-lived messenger produced in the cells. This messenger plays a significant role in regulating vasodilation under hypoxia.
The process by which nitrogen monoxide binds to myoglobin is already well characterized experimentally, which is important for the calibration of computer simulations. There is also experimental evidence for the existence of metastable intermediates, but our simulations provide insights into the underlying chemical structure and the dynamics of these intermediates, and thus the function of the protein.
Meuwly
Experimental observations and computer simulations are thus both important for understanding complex biological and chemical systems. This combination will indeed provide answers to questions related to the optimization and adaptation of proteins or pharmaceutical agents that are active. This highlights the need to understand the procedures at the atomic and molecular level.