Summary
Characterization of exosomes has become a topic of interest in medical research and molecular biology. This paper contains a summary of how the Wyatt Eclipse® Field-Flow Fractionation system coupled to multi-angle light scattering (MALS) and dynamic light scattering (DLS) equipment allows a more detailed and complete characterization of exosomes.
This is done through a physical separation of exosomes from other components of the biological matrix, such as serum, urine, etc., followed by online determination of charge, size, and composition. Fractions can be separated for further off-line analysis, in particular, sequencing of the DNA contained within exosomes.
Recently in Nature Biology, a publication by Zhang et al. has shown that biological function corresponds with size, and three distinct size populations could be separated for the first time using the Wyatt Eclipse FFF system. This article explains how the separation of exosomes can be achieved, and what information is obtained by combining DLS and MALS online.
Introduction
Exosomes are a unique source of specific disease biomarkers, according to recent studies. Non-affected and disease-affected cells with a size range of 30-100 nm release exosomes, small vesicles, as do all other cells. Microvesicles, however, have a usual diameter of 100-1000 nm, and release from cells of origin in a different way.
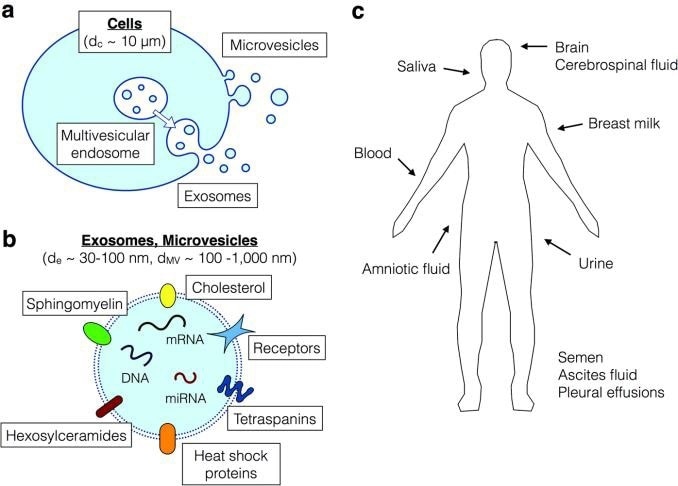
Figure 1. Secretion of exosomes from cells and their significance for biomarkers. a) biogenesis of exosomes from multivesicular endo-somes, in contrast to the microvesicles which are released from the cell surface via membrane budding. b) exosomes and microvesicles both contain transmembrane proteins, intracellular proteins, DNA, RNA and miRNA, which are potential biomarkers. c) exosomes are found in all body fluids. Figure from (2).
Exosomes originate from active cells, such as active tumor cells, whereas microvesicles come primarily from blebbing of dying cells and do not reflect the active disease status, thus are of lesser interest with respect to biomarkers. Exosomes possess a double lipid layer containing DNA, RNA, miRNA, soluble and transmembrane proteins, reflecting the biological repertoire of the cell.
To use exosomes as a source of biomarkers, they must be separated from microvesicles and soluble high abundance proteins. Bulk exosomes are commonly collected using non-specific methods such as ultracentrifugation or precipitation.
FFF is the method of choice, as it is highly effective at size separation within 1-1000 nm. Zhang et al. has demonstrated the unique capability of FFF in isolating exosomes from other material, and separating the exosomes into three different populations with distinct properties and biomarkers.
How Field-Flow Fractionation Works
The sample is injected into a thin ribbon-like channel with a porous bottom wall. Some of the flow passing through the channel towards the outlet permeates through the porous bottom, creating a cross-flow. Friction force concentrates the sample toward the accumulation wall due to the cross-flow, against which Brownian motion acts as a counterforce.
The molecules are constantly moving away from the bottom wall, but are pushed back from above. This creates a particle cloud with distance from the bottom wall exponentially decreasing the concentration, as when air pressure decreases with distance above sea-level. Particles of different size will have a different median distance from the accumulation wall, given by its diffusion coefficient.
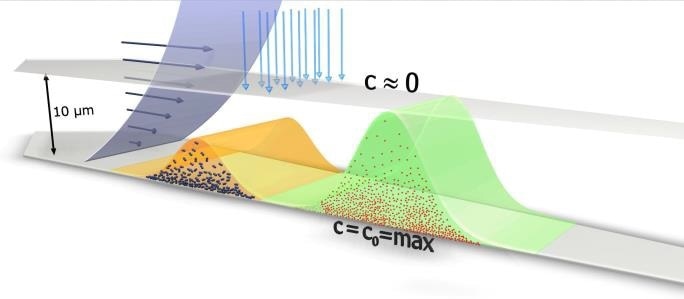
Figure 2. Principle of Flow-FFF separation. Shown are two particle clouds moving along the channel accumulation wall, which consists of a membrane supported by a frit. The cross flow is coming from the top and permeates through the accumulation wall exerting a friction force on the particles. The smaller (red) particles diffuse higher up against the cross flow compared to the bigger (blue) particles. The parabolic velocity flow profile of the channel flow transports the more extended particle cloud much faster, compared to the more compact particle cloud of the bigger particles.
Smaller particles possess a greater diffusion coefficient and so will be higher up in the channel, and larger particles vice versa. The distance between the area of maximum concentration near the accumulation wall, and the channel with no molecules present is less than 10 µm. The average height of the channel is 350 µm, meaning that the sample stays close to the bottom wall throughout the process of separation.
Smaller particles move more quickly through the channel towards the outlet, as the laminar flow velocity becomes greater with increasing distance from the accumulation wall. Thus, particles are separated by size, with smaller particles eluting more quickly (Figure 2). FFF theory is well explained, giving an equation that describes retention in a quantitative way, as given below.
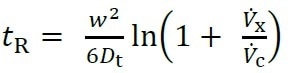
Where Dt is the diffusion coefficient, Vx and Vc are the cross-flow and channel flow rates, respectively, tR is the measured retention time, and w is the height of the channel.
SCOUT DPS® software predicts the results of an experiment based on this equation.
Materials and Methods
Separation of exosomes was done using an Eclipse AF4 system with an Agilent 1260 HPLC equipped with a fraction collector. A DAWN® with WyattQELS™ installed at detector 12 was used for MALS and DLS detection. Instrument control was performed using the Eclipse Plug-in for ChemStation. A detailed experimental protocol is described in a separate protocol exchange paper.[4] Separation was achieved using a short channel and a standard flow program, gradually lowering the cross-flow from 0.5 ml/minute to 0 in 45 minutes.
Data Processing
Data processing was done using ASTRA®, which has the unique ability to combine both Multi-Angle Light Scattering (MALS) and dynamic light scattering (DLS) in a single package.
Why MALS and DLS Combined?
Both light scattering techniques measure nanoparticle size, and are highly complementary. MALS determines rms radius from 10 nm to 500 nm, and is about 20x more sensitive than DLS, while DLS determines hydrodynamic radius between 0.5 nm - 200 nm, with the upper limit being determined by the flow-rate. The combination of MALS and DLS is indicative of shape and structure.
An optional DLS module can be incorporated into DAWN, utilizing the same flow cell and laser, making it extremely versatile.
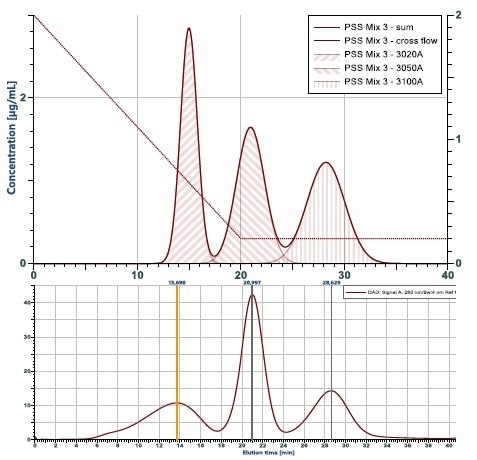
Figure 3. Simulation of a fractionation compared to the experi-mental outcome. Shown is the theoretical calculation of a fractogram from a mix of three particles with 10, 20 and 50 nm radius. Below is the experimental results. It comes very close to the prediction, with the exception of the wider peak for the 10 nm particle eluting first. The reason is, that this particle is not monodisperse as assumed in the theoretical calculation.
Results and Discussion
In their Nature Cell Biology paper "Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric-flow field-flow fractionation", Zhang et al. found that all exosomes do not have the same essential nature.
Using FFF-MALS-DLS, they identified three distinct types of small extracellular vesicles, termed Exo-L, Exo-S and Exomeres. Exo-L are large exosome vesicles of between 90 - 120 nm in diameter, Exo-S are smaller exosome vesicles with diameters between 60 – 80 nm, while exomeres are non-membrane bearing particles of around 35 nm in diameter.
Other biophysical properties in addition to size and presence of a membrane distinguish these types, including degree of N-glycosylation, protein content, metabolite content, DNA/RNA profiles, and function. However, these types are conserved across most cell lines, suggesting that they are independent classifications of bio nanoparticle with distinct origins and function.
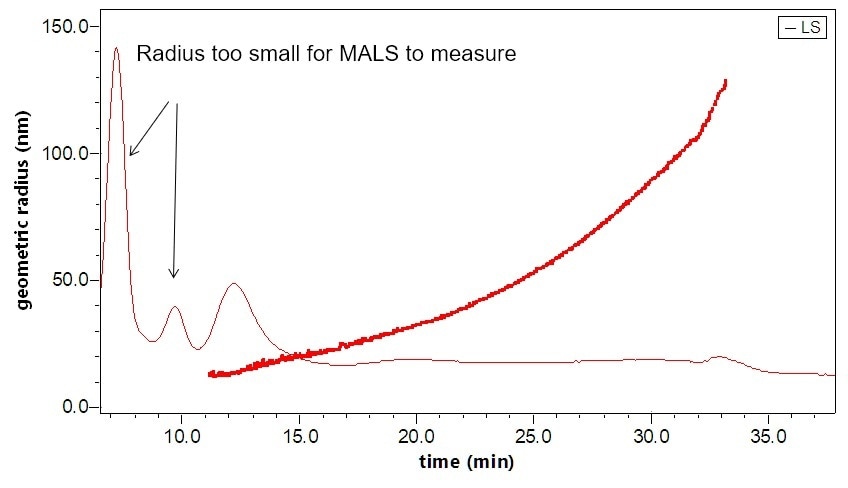
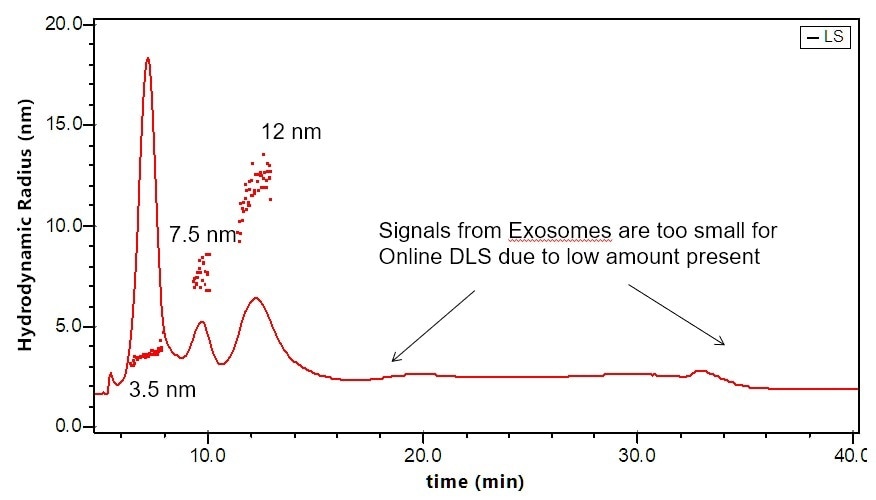
Figure 4. Separation and characterization of exosomes by FFF-MALS-DLS. The triple combination allows for unique characterization and understanding of these bionanoparticles. Utilizing MALS and DLS in tandem extends the range of size measurements, which has proven critical based on recent published results.
Conclusions and Outlook
FFF-MALS-DLS is a diverse, accurate, and powerful platform for separating and characterizing exosomes and other nanoparticles. It supports online detectors such as UV/Vis, fluorescence, absorption, mass spectrometry, differential refractometry via Optilab®, in addition to light scattering detectors.
Electrical asymmetric-flow field-flow fractionation (EAF4), a new technology, uses the Mobility™ system to separate particles based on size and charge. The Mobility system can also determine zeta potential and so provide full, detailed distributions of size vs. zeta potential.
The Zhang paper is not the first to make use of the Eclipse/DAWN system, as the same setup had previously been employed by Yang et al. to identify a potentially life-saving biomarker, that there is a significant difference in size between urinary exosomes from healthy people compared with patients suffering from prostate cancer. Other publications making use of FFF-MALS-DLS to characterize exosomes have made similarly important discoveries.

References
- Zhang H. et al. (2018) "Identification of distinct nanoparticles and subsets of extracellular vesicles by asymmetric flow field-flow fractionation", Nat. Cell Bio. 20, pp. 332–343 DOI: 10.1038/s41556-018-0040-4
- Sitar S. et al. (2015) "Size Characterization and Quantification of Exosomes by Asymmetrical-Flow Field-Flow Fractionation", Anal. Chem. 87, pp. 9225-9233 DOI: 10.1021/acs.analchem.5b01636
- Petersen K.E. et al. (2014) "A review of exosome separation techniques and characterization of B16-F10 mouse melanoma exosomes with AF4-UV-MALS-DLSTEM", Anal. Bioanal. Chem. 406, pp. 7855-7866 DOI: 10.1007/s00216-014-8040-0
- Zhang H. and Lyden D. (2018) "A protocol for Asymmetric-Flow Field-Flow Fractionation (AF4) of small extracellular vesicles", Protocol Exchange, 19 February 2018, DOI: 10.1038/protex.2018.002
- Yang J.S. et al. (2017) "Size Dependent Lipidomic Analysis of Urinary Exosomes from Patients with Prostate Cancer by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography-Tandem Mass Spectrometry", Anal. Chem. 89, pp. 2488-2496 DOI: 10.1021/acs.analchem.6b04634
- Agarwal K. et al. (2015) "Analysis of Exosome Release as a Cellular Response to MAPK Pathway Inhibition", Langmuir 31(19), pp. 5440-5448 DOI: 10.1021/acs.langmuir.5b00095
- Ashames A. (2015) "Development of separation methods to produce uniform exosomes subpopulations using field-flow fractionation techniques", Ph.D. thesis. Colorado School of Mines DOI: http://hdl.handle.net/11124/17052
- Kang D. et al. (2008) "Proteomic Analysis of Exosomes from Human Neural Stem Cells by Flow Field-Flow Fractionation and Nanoflow Liquid Chromatography Tandem Mass Spectrometry", J. Proteome Res. 7 pp. 3475-3480 DOI: 10.1021/pr800225z

This information has been sourced, reviewed and adapted from materials provided by Wyatt Technology.
For more information on this source, please visit Wyatt Technology.