Electron Diffraction, also known as 3DED/microED (3-Dimensional Electron Diffraction or micro Electron Diffraction), is a revolutionary analytical technique for the advancement of structural science.

Image Credit: Rigaku Corporation
Single-crystal X-ray diffraction (SC-XRD) is often referred to as the gold standard of sample characterisation, offering three-dimensional structure of both molecules and their crystalline packing, typically with high precision. Using current state-of-the-art X-ray diffractometers, measurement of samples down to about 1 micron is possible. With the advent of electron diffraction, nanocrystals below this limit, even down to tens of nanometres, can also be elucidated.
Prior to electron diffraction becoming available, synthetic chemists were forced to use NMR to try to postulate the 3D structure of new materials that only formed nanocrystals. For complex molecules, NMR is extremely challenging to interpret. As a result, researchers looked for alternatives.
Early works published by Kolb et al. (2007) and Shi et. al. (2013) paved the way for the development of this new technique.
Evolution of the World’s First Electron Diffractometer
Recognizing the potential of electron diffraction for the crystallography community, Rigaku and JEOL embarked on a partnership to develop the world’s first commercially available electron diffraction system in 2020.
The following year, the collaboration culminated in the XtaLAB Synergy-ED, a dedicated electron diffractometer. The XtaLAB Synergy-ED fully integrates expertise from both companies, i.e.:
- Rigaku’s high-speed, high-sensitivity detector (HyPix-ED) and high-powered crystallographic software
- JEOL’s expertise in the generation and control of stable electron beams
The system incorporates a seamless workflow from data collection to structure determination of three-dimensional molecular structures using CrysAlisPro-ED, an extension of Rigaku’s proven CrysAlisPro structural determination software for SC-XRD.
Electron diffraction differs from SC-XRD in that it uses electrons as the source rather than X-Rays. This enables crystallographers to examine crystals smaller than the ~1 µm limit of SC-XRD.
Due to the fact that electrons interact with matter to a much greater extent compared to X-Rays, smaller crystals — of the order of tens of nanometers — are ideally suited to electron diffraction. This makes it a highly complementary technique to SC-XRD.
Challenges for Electron Diffraction
The challenges for electron diffraction are different from those of SC-XRD
- Absorption – Small crystals are often better, as even small crystals can completely stop the electron beam.
- Dynamical Diffraction – Electron beams are diffracted multiple times as they traverse through the crystal. Therefore, the thicker the crystal, the greater the dynamical effect, which can be problematic for resolving and refining crystal structures.
- Sample decay – Exposing crystals to high-energy electron beams result in decay. This can be overcome by using low-temperature data collection and/or a carefully selected beam intensity and scan speed.
Electron Diffraction Systems

Image Credit: Rigaku Corporation
Rigaku’s XtaLAB Synergy-ED consists of a vertical vacuum column that ensures the sample stays in the beam throughout the scan. The column incorporates a JEOL electron source and optical system.
Samples are mounted on a stage that allows x, y, z and tilt adjustment for precise sample positioning. A cryo-transfer sample holder is also available for sensitive samples.
The XtaLAB Synergy-ED uses a HyPix-ED detector. This is similar to other HyPix detectors used in SC-XRD in that it is event-driven, meaning it:
- Counts each diffracted electron
- Does not produce any dark noise or readout noise
- Offers high frame rates
- Produces high-quality data
The system is controlled using the same CrysAlisPro software package as Rigaku’s SC-XRD systems which means anyone familiar with those systems will be able to perform electron diffraction experiments almost immediately. The only difference is the sample visualizing and centering functions which are self-explanatory and intuitive. The analytical workflow is identical.
As with X-Ray diffraction experiments, CrysAlisPro performs data acquisition and data processing concurrently. AutoChem, which is fully integrated into CrysAlisPro, performs fast, fully automated structure solution and refinement during data collection, thereby optimizing productivity.
Due to the mounting approach and limited sample access in electron diffraction experiments, it is often necessary to merge data from multiple grains to generate a complete dataset. CrysAlisPro makes this a simple operation, also applying all corrections and scaling to produce the final hkl reflection file.
Sample Preparation
Preparing samples for MicroED experiments is relatively straightforward, involving:
- Preparing fine-grained samples
- Attaching to TEM grids
- Loading grids into the XtaLAB Synergy-ED
Results
Below are examples of structures that have been determined using electron diffraction, which typically allows you to see hydrogen positions in the different maps. All samples were examined using the XtaLAB Synergy-ED using the CrysAlisPro software. Structures were refined automatically by AutoChem or manually using Olex2.
Structures were refined using SFAC instructions suitable for electron diffraction. In some instances, single grains provided sufficient completeness for the structural solution. In other cases, structures were resolved by merging datasets from multiple grains.
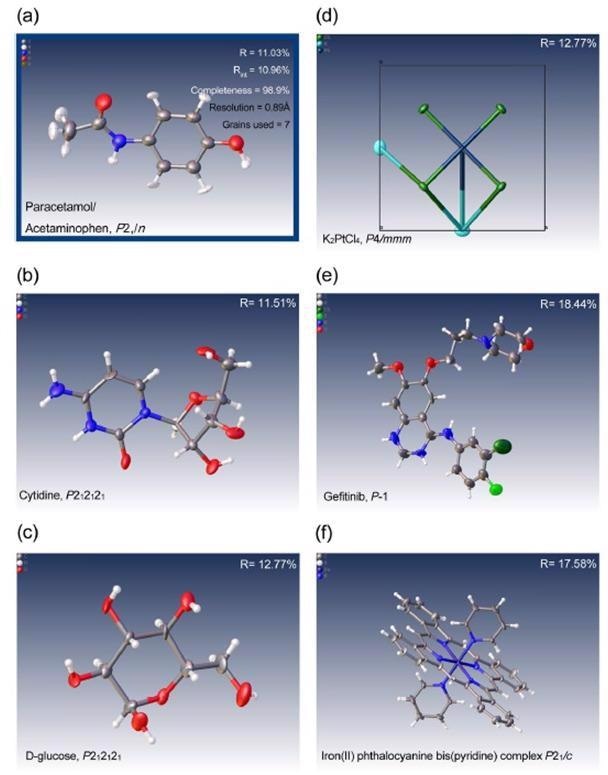
Some examples of the first attempts at solving structures with data collected on the XtaLAB Synergy-ED including pharmaceutical compounds, inorganic minerals and complexes ranging from triclinic to tetragaonl symmetry. (a) paracetamol (acetaminophen), (b) cytidine, C0) glucose, (d) potassium tetrachloroplatinate, (e) gefitnib and (f) iron(II) phthalocyanine bis(pyridine) complex. Image Credit: Rigaku Corporation
The first published structures using data from an XtaLAB Synergy-ED system were reported by researchers from the University of Birmingham and the University of Nottingham in the journal Nature Communications.
In this paper, a research team, led by Professor Neil Champness, successfully determined the structure of the difficult rotaxane crystals using electron diffraction. This followed unsuccessful attempts to resolve the structure using X-Rays on a synchrotron beamline.
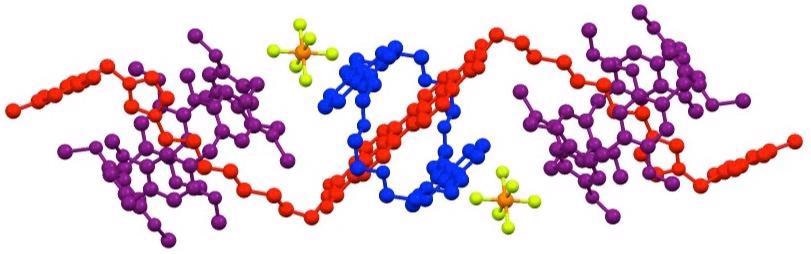
Rotaxane structure determined using the Rigaku XtaLAB Synergy-ED electron diffractometer. Image Credit: Rigaku Corporation
Applications
This technique has the potential application in fields such as:
- Catalysis
- Energy storage and alternative energies
- Aerosol research
- CO2 reduction
- Biomineralization
- Pharmaceuticals
- Medical technology
- Mineralogy
- Archaeology
Conclusion
Direct structural determination of nanocrystals and samples too small for conventional X-Ray diffraction studies is now possible using electron diffraction.
For the first time, a totally dedicated electron diffractometer is commercially available in the XtaLAB Synergy-ED, which is able to produce structural refinements from previously unmeasurable samples.
This turnkey system is the ideal complement to existing single crystal diffractometers and, due to its compact design, can be easily accommodated into existing X-Ray labs.
Driven by the CrysAlisPro software interface, which is familiar to many crystallographers, users will be able to perform electron diffraction experiments with almost no training, with the potential to rapidly generate publications using this emerging analytical technique.
References
- Kolb et al, (2007), Towards automated diffraction tomography: Part I—Data acquisition, Ultramicroscopy, 107, (6-7), 507-513
- Shi et al. (2013), Three-dimensional electron crystallography of protein microcrystals, eLife
- Pearce et al., (2021), Selective photoinduced charge separation in perylenediimide-pillar[5]arene rotaxanes, Nature Communications 13, 415 (2022). https://doi.org/10.1038/s41467-022-28022-3

This information has been sourced, reviewed and adapted from materials provided by Rigaku Corporation.
For more information on this source, please visit Rigaku Corporation.