There have been significant developments in recent years regarding thermal treatments, which are beginning to replace traditional techniques in the preservation of highly perishable products. This has taken the form of new thermal (e.g., ohmic heating, microwaves) and non-thermal (e.g., high-pressure, cold-plasma) treatments, which have proven to be less damaging to the products (Hassoun et al., 2020).
These treatments, among other benefits, allow for an additive-free extension of food shelf life, particularly unsaturated fatty acids, micronutrients (e.g., amino acids and vitamins) and aromatic compounds.
When it comes to inactivating microorganisms and enzyme activity in many food products without compromising the quality of their sensory attributes, high-pressure (HP) treatments are recommended.
For meat products or fish in particular, however, HP treatment favors certain changes that result in an increase in hardness, a decrease in volume, a change in color, and a disruption of hydrophobic and electrostatic interactions, which lead to changes in secondary, tertiary and quaternary structures (Jolvis Pou, 2021; Oliveira et al., 2017).
Depending on the process temperature, as well as the pressure level and duration, the conformation of proteins can be modified, leading to aggregation, denaturation or gelation (Messens et al., 1997).
Fresh fish muscles are particularly sensitive to HP treatment because they are composed of about 20% protein. The main proteins in fish muscles include myofibrillar proteins (40–60%), sarcoplasmic proteins (30%), and stromal proteins (i.e., collagen) (Truong et al., 2015).
Fourier transform infrared spectroscopy (FTIR) analysis can be used to study changes in the conformational structure of myofibrillar proteins (Cando et al., 2014).
FTIR spectroscopy is widely considered an established, rapid, and direct method to detect, identify and evaluate the vibrations of specific bonds and functional groups attributed to a characteristic molecular structure.
This method also offers several advantages, including the ability to simultaneously evaluate overall sample composition and analyze small sample quantities, with optional simple sample pretreatment and no need for wet chemistry.
The basis of many applications of FTIR spectroscopy in food analysis is the analysis of the fingerprint region (400–1800 cm-1). The region between 1600 and 1700 cm-1, in particular, corresponds to the amide I band, providing insight into changes in the secondary structure of proteins.
The changes in the secondary structure of myofibrillar proteins in salmon samples were investigated by this study when subjected to different levels of high pressure.
Materials & Methods
One day after their harvest date, twelve whole salmon (Salmo salar, farmed in Scotland) were purchased from a local supplier. A slice roughly 5 cm wide was excised from each fish and then cut into four parts (about 40 g each). Each of the samples was then vacuum-packed into an individual polyamide/polyethylene bag and kept on ice before being processed with high pressure.
A custom-made high-pressure device was used, into which the packed samples were inserted via the stainless-steel chamber.
Equipped with a temperature regulator device and water jacket, the chamber was filled with water at a temperature of 20 °C. Pressure was generated by the addition of water into the chamber using a hydrostatic pump. One sample per fish was kept unpressurized for a control.
For a duration of 5 minutes, test samples were processed with a balanced experimental design for 5 minutes at 200 MPa, 400 MPa, and 600 MPa, with a compression rate of 3 MPa/second. The depression rates were respectively 4 MPa/seconds, 8 MPa/second, and 10 MPa/second, whilst the maximal temperatures reached were 24.7 °C, 29.4 °C, and 33.6 °C.
The samples were kept on ice as soon as the HP treatment ended. Before being cooled with liquid nitrogen (–196 °C), each sample cube was plunged and stirred for 30 seconds in 2-methylbutane (–160 °C). Until analysis, the cryofixed samples were then kept at –80 °C.
The cryofixed muscle sample was used to obtain histological cross-sections (6 μm thick), which were then collected on a Barium Fluoride window that was compatible with IR spectroscopy, and subsequently air-dried at room temperature.
Dehydrated samples were preferable to avoid the intense signal from water in infrared, which would superimpose on the signal of the proteins’ amide I band at ~1650 cm-1.
The Thermo Scientific™ Nicolet™ iN10 Microscope was used to collect infrared spectra scanning from 4000 to 675 cm-1 with a spectral resolution of 4 cm-1 and an aperture size set at 30x30 μm. At least 20 spectra were acquired for each section in 20 different muscle fibers. The result of 64 accumulated scans is what produced each spectrum.
By accumulating 128 scans, the accumulated spectra were averaged and deducted from a background spectrum obtained at the start of the scan.
Thermo Scientific TQ Analyst™ Pro Edition Software was used to analyze the acquired spectra. A multiplicative signal correction (MSC) was first applied to all spectra to remove unwanted interference. Then, a second derivative was calculated for all spectra associated with a Saviztky-Golay filter with nine points and a third-degree polynomial.
The analysis was primarily focused on the 1600–1700 cm-1 region, which is mostly assigned to the amide I band of proteins. On all spectra, principal component analysis (PCA) was performed using the absorbency at each measured wave number as a variable to explore similarities between the samples without prior assumptions.
To determine which variables had the most weight to describe the variability between the samples and the pressure conditions, principal component representation was used.
Results and Discussion
An average FTIR spectrum was shown in Figure 1 of between 4000 and 675 cm-1 derived from spectra of the untreated raw salmon muscle fibers. For all the samples investigated, this spectrum represents the most typical features seen.
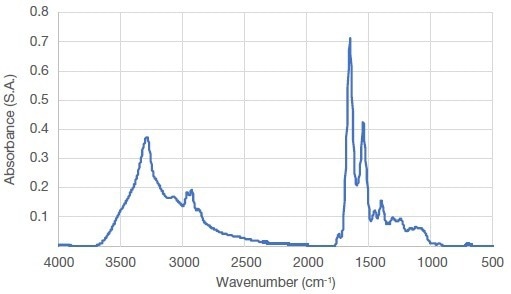
Figure 1. Average FTIR spectrum of raw salmon muscle. Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The amide A band (~3100– 3600 cm-1) is attributed principally to OH and NH stretching vibrations. A typical band attributed to lipids and ester groups is observed at ~1750 cm-1. Amide I, II, and III can be found at ~1650, 1550, and 1260 cm-1, respectively. Finally, depending on the side chain, backbone stretching vibrations contributed to a number of modes in the 1180–920 cm-1 region.
The specific secondary structures of proteins can be determined by the study of changes in the amide I band, often presented as a useful tool. The amide I band originates mainly from C=O is located in the region between 1600–1700 cm-1, and is weakly coupled to N-H in-plane bending and C-N stretching vibrations.
The amide I region (1700–1600 cm-1) was then studied as a second derivative spectrum to better understand the changes related to the secondary structure of myofibrillar proteins (Figure 2).
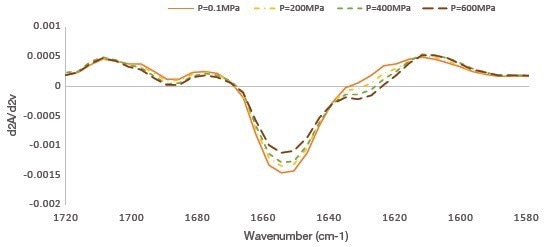
Figure 2. Second-derivative average FTIR spectra (1700–1600 cm-1) obtained in raw salmon muscle of unpressurized (0.1 MPa) and pressurized (200, 400, and 600 MPa). Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
The second derivative spectra clearly showed a decrease in absorbency at 1655 cm-1 and an increase in absorbency at 1688 and 1628 cm-1 with increasing pressure.
The central band at 1655 cm-1 corresponds to α-helical structures, while the band components around 1688 cm-1 and below 1640 cm-1 are typically associated with intramolecular, antiparallel β-sheet structures, and those in the range between 1610 and 1630 cm-1 with intermolecular, aggregated, antiparallel β-sheet structures (Barth & Zscherp, 2002).
The researchers used the height of the peak of the second derivatives for the bands located at 1655 cm−1 (α-helix) and 1628 cm-1 (β-sheet) to underline the evolution of the muscle fibers’ secondary structure.
Figure 3 presents the results as histograms. At a pressure of 200 MPa, a change in the secondary structure of the salmon muscles was observed, and at higher levels of pressure this modification of the secondary structure was enhanced.
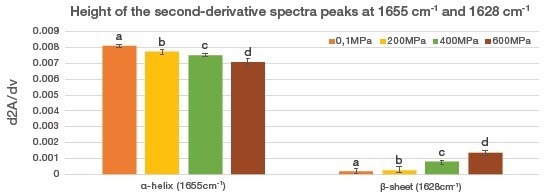
Figure 3. Evolution of α-helixes and aggregated β-sheets in salmon measured by the height of the second-derivative spectra peaks at 1655 cm-1 and 1628 cm-1, respectively. The results are expressed as the mean ± standard error. Means with different letters are significantly different (P < 0.05). Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
By unfolding myofibrillar proteins and altering the properties of hydrogen bonds, HP treatments could therefore generate destabilization of non-covalent interactions in the secondary and tertiary structures, particularly interactions that are ionic and hydrophobic.
As was shown by PCA performed on all the samples, the four pressure groups are clearly separated along the first principal component (control versus 200 MPa, 400 MPa, and 600 MPa), explaining 65.22% of the total variance (Figure 4).
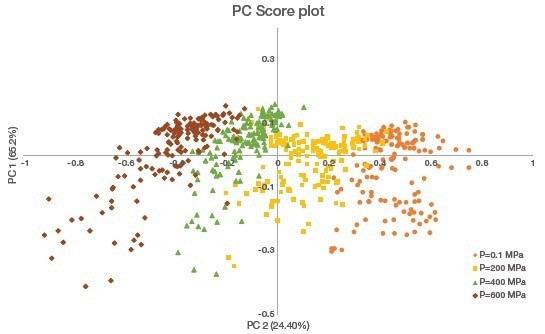
Figure 4. PCA of the second derivative of the amide I (1600–1700 cm-1) band. Pressure groups are separated along the first principal component (PC1). Image Credit: Thermo Fisher Scientific – Materials & Structural Analysis
Though data is not shown, the first axis of the principal component was positively oriented for the band at 1655 cm-1 and negatively oriented for the band at 1628 cm-1. These orientations indicate that increasing pressure decreased α-helical structures and increased β-sheet structures within salmon myofibrillar proteins.
Conclusions
The use of FTIR enabled the direct characterization of the secondary structure of myofibrillar proteins on sections of muscle tissue that had not been chemically treated and thus preserved their molecular assemblies.
The study uncovered significant variations in the overall degree and nature of protein aggregation under high pressure, especially when performed above 200 MPa. These secondary structure changes can explain observed modifications, including a color change and an increase in hardness after high-pressure treatments.
Understanding the effects of high pressure at the molecular level could thus help researchers comprehend the changes in fish quality and establish the best treatment parameters to reduce the undesirable impacts of HP treatments.
Acknowledgments
Special thanks to Camille Renaud, Marie de Lamballerie, Laurence Pottier, Claire Guyon, Thierry Astruc, and Annie Venien, who allowed us to extract their FTIR results from the study, “Effects of high-pressure treatment on the muscle structure of Salmon (Salmo salar)” (Renaud et al., 2022). This study was funded by Pays de la Loire as part of the PATACHON project.
References
- Barth A., Zscherp C. (2002) What vibrations tell us about proteins. Q Rev Biophys 4: 369–430. https://doi.org/10.1017/S0033583502003815
- Cando D., Moreno H. M., Tovar C. A. et al. (2014) Effect of high pressure and/or temperature over gelation of isolated hake myofibrils. Food Bioproc Tech 7(11): 3197–3207. https://doi.org/10.1007/s11947-014-1279-9
- Hassoun A., Ojha S., Tiwari B. et al. (2020) Monitoring thermal and non-thermal treatments during processing of muscle foods: a comprehensive review of recent technological advances. Appl Sci 10: 6802.
- Jolvis Pou K. R. (2021) Applications of high pressure technology in food processing. Int J Food Stud 10(1): 248–281. https://doi.org/10.7455/IJFS/10.1.2021.A10
- Messens W., van Camp J., Huyghebaert A. (1997) The use of high pressure to modify the functionality of food proteins. Trends Food Sci Technol, 8(4): 107–112. https://doi.org/10.1016/S0924-2244(97)01015-7
- Oliveira F. A., de Neto O. C., Santos L. M. R. et al. (2017). Effect of high pressure on fish meat quality – A review. Trends Food Sci and Technol 66: 1–19. https://doi.org/10.1016/j.tifs.2017.04.014
- Renaud C., de Lamballerie M., Guyon C. et al. (2022). Effects of high-pressure treatment on the muscle structure of salmon (Salmo salar). Food Chem 367: 130721. https://doi.org/10.1016/j.foodchem.2021.130721
- Truong B. Q., Buckow R., Stathopoulos C. E., Nguyen M. H. (2015). Advances in high-pressure processing of fish muscles. Food Eng Rev 7(2): 109–129. https://doi.org/10.1007/s12393-014-9084-9

This information has been sourced, reviewed and adapted from materials provided by Thermo Fisher Scientific – Materials & Structural Analysis.
For more information on this source, please visit Thermo Fisher Scientific – Materials & Structural Analysis.