The ‘SRRF-Stream’ is a new real-time super-resolution microscopy functionality that works entirely on Andor’s iXon Life and iXon Ultra EMCCD cameras.
SRRF-Stream Super-Resolution
- Real time – improved workflow, avoids post-processing. View in ‘Live Mode’
- Live cell dynamics – full FOV super-res images every 1-2 seconds. > 10 fps using ROI
- Low excitation intensities (mW-W/cm2) – prolonged live cell observations and accurate physiology
- Conventional fluorophores, for example, GFP – simple labeling, no photo-switching necessary
- Cost-effective – convert conventional fluorescence microscopes to super-resolution microscopes
iXon SRRF-Stream Comparison
Mitochondria
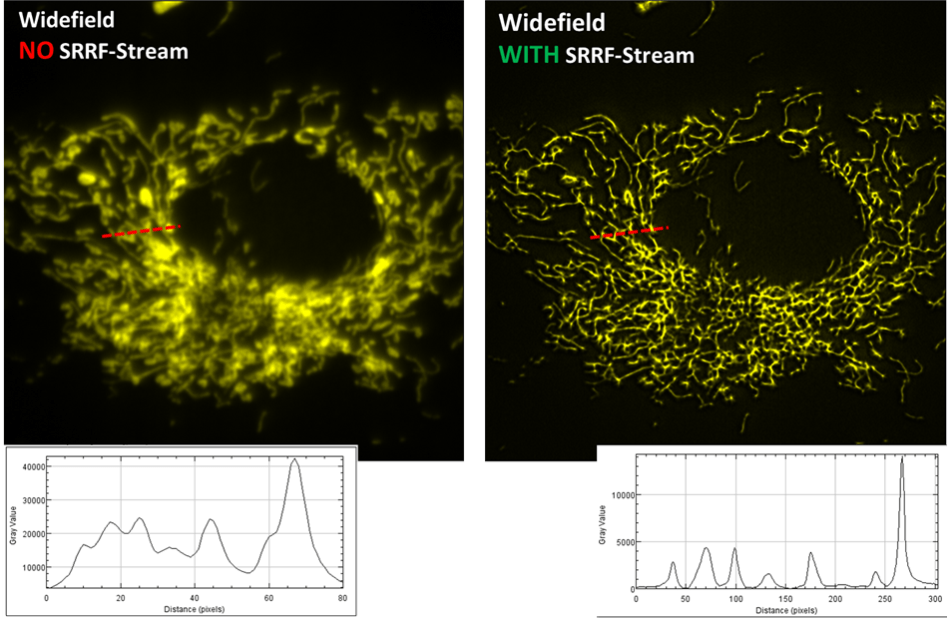
Image comparison of a fluorescently labeled BPAE cell, recorded with a widefield fluorescence microscope and a SRRF-Stream enabled iXon Life 888 EMCCD camera. A x63 objective was used, with further 2x magnification and 560 nm illumination. 100 raw ‘input’ images were recorded for every resultant super-resolution image, resulting in a super-resolution image rate of 0.5 Hz. For an impartial comparison without SRRF-Stream, 100 standard widefield images were recorded and then averaged. While the original image was of a larger field of cells, a zoomed ROI of one cell is illustrated here so as to more easily exhibit a line intensity profile comparison through a small region. The enhancement in resolving power is readily obvious.
Mitotic Division
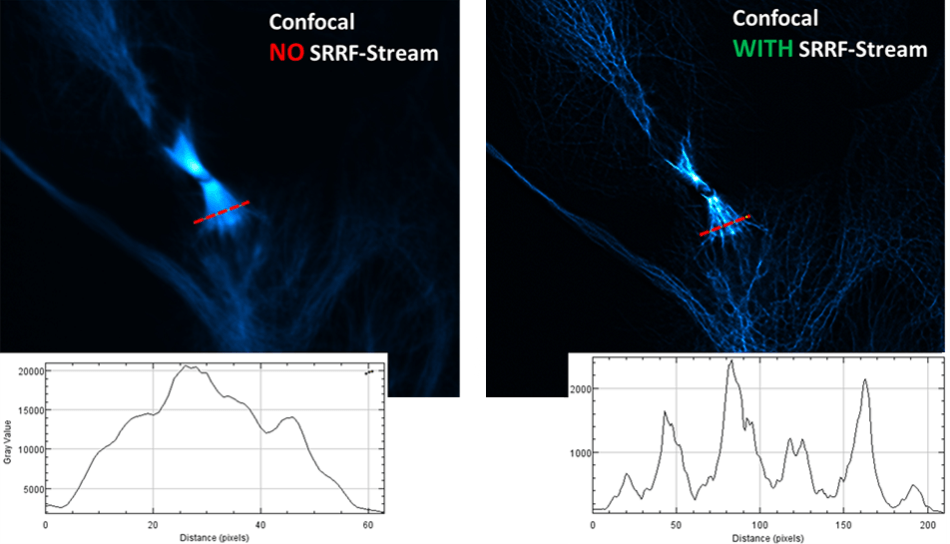
This image comparison of a fluorescently labeled U2OS cell line* was recorded with an Andor Dragonfly confocal spinning disk fluorescence microscope and a SRRF-Stream enabled iXon Life 888 EMCCD camera. A x63 objective was used, with additional 2x magnification and 488 nm illumination. An unparalleled improvement in resolving power can be noticed in the level of detail in the mitotic spindle. This is additionally evidenced in the comparative line intensity profile drawn through this region.
*U2OS cell line was fixed, stained with anti-alpha-tubulin primary antibody (green, AF488) and phalloidin (red, rhodamine) to visualize F-actin, DAPI staining to visualize nuclei. Samples prepared by Klebanovych A., Laboratory of Biology of Cytoskeleton, IMG of the AS CR, v.v.i.
SRRF vs SIM
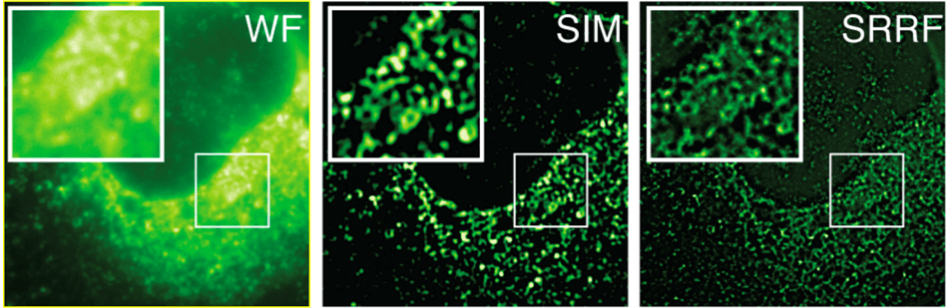
HCV infected cells stained with anti-NS5A. Here, a comparison of Widefield (WF), Structured Illumination Microscopy (SIM) and SRRF images (SRRF of the widefield image) are done. The images are of the same field of cells, recorded on the same microscope, using identical objective and optical path. The sole difference being that SIM was recorded using an sCMOS detector with 6.5 mm pixels while the Widefield and resultant SRRF was recorded using an iXon EMCCD detector with 16 mm pixels. The greater resolving power of SRRF is evident, indicative that SRRF is accomplishing a greater than two-fold improvement over the classical diffraction limit. SIM is theoretically limited to a two-fold reduction of the classical diffraction limit. Sample courtesy of the Grove lab at UCL.
Mitotic Division 2
Mammalian cell experiencing mitoses. Blue signifies DNA staining, Green microtubules and Red kinetochores. The left image illustrates a widefield z-stack and right image the equivalent acquired with SRRF-Stream. Sample courtesy of Phil Auckland at Warwick University, imaging by the Henriques laboratory at University College London (UCL).
BCS-40 Membrane
200s time-lapse of a live BSC-40 cell labeled with cell mask and imaged with 635 nm LED illumination. The first 100 frames correspond to widefield imaging with a 1 second exposure; the second 100 frames correspond to SRRF-Stream imaging where each frame is produced from SRRF-Stream processing of 50 images (20 ms exposure time). Sample preparation by David Albrecht (Ricardo Henriques and Jason Mercer labs at UCL).
Clathrin Pits
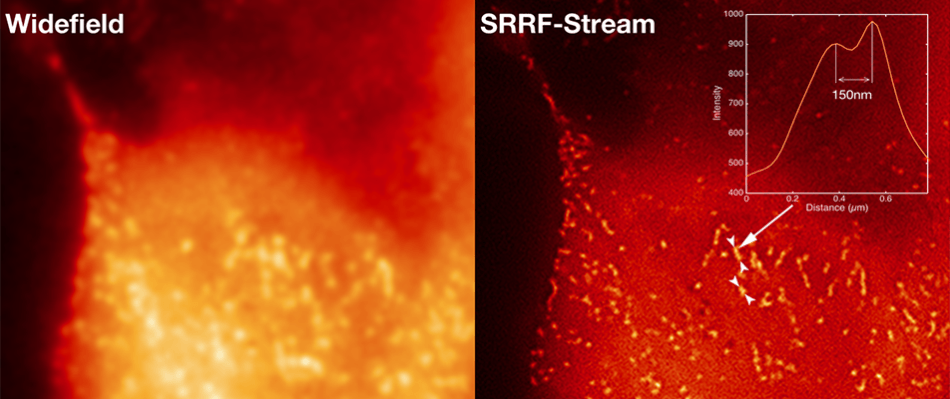
Comparative images of Clathrin coated pits of live HeLa cells, labeled with mCherry, recorded on a widefield microscope at 2 FPS. 100 raw ‘input’ images were recorded for every resultant super-resolution image, resulting in a super-resolution image rate of 2 FPS. A line intensity profile is shown through a small region of the SRRF-Stream image, representing resolution of structures that are 150 nm apart. Sample preparation by Caron Jacobs (Ricardo Henriques and Mark Marsh labs at UCL).
Yeast
Comparative 3D projection montages of fission yeast lifeAct expressing strain. Recorded with standard widefield versus SRRF-Stream widefield, using identical exposure times. Strain courtesy of Mohan Balasubramanian’s laboratory (U. Warwick) Sample courtesy of Gautam Dey (Buzz Baum laboratory at UCL).
Blood
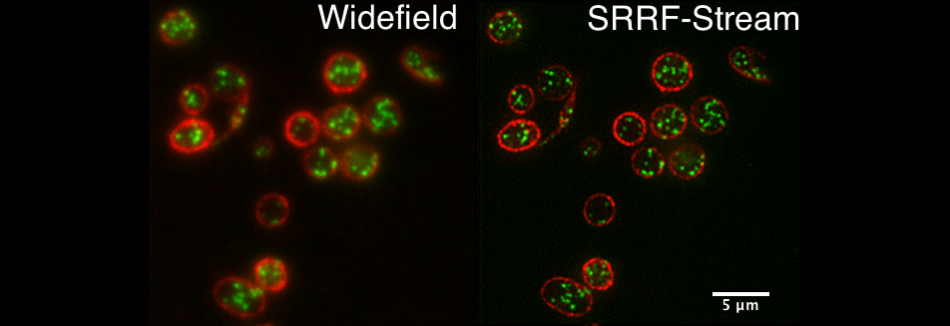
Comparative standard widefield and widefield SRRF-Stream images of blood platelets, red membrane, green internal granules. Sample courtesy of Cutler laboratory at UCL.
Tubulin
A still widefield image of a live HeLa cell expressing tubulin-GFP followed by a SRRF-Stream time-lapse of the same region at 1fps (SRRF-Stream analysis of 50 frames at 20 ms exposure). Sample preparation by David Albrecht (Ricardo Henriques and Jason Mercer labs at UCL).
iXon SRRF-Stream Applications
With its ability to push through the classical diffraction limit, and additionally, to realize this in real time, with non-complex sample labeling, conventional equipment and with low intensity illumination, the SRRF-Stream makes way to unlock formerly unobserved cellular structure and behavior, at unparalleled spatio-temporal resolution in a low photo damage friendly manner.
Observational Capabilities of SRRF-Stream
- Elucidation of protein structure analysis at a sub-organelle level
- Tracking of individual molecules inside cells
- Using this tracking to gain understanding into separate molecular machinery underpinning cellular physiology
- With this new information, update models of cell function
Applications of SRRF-Stream
- Membrane fusion involving separate SNARE protein machinery
- Dynamic variations of and within synaptic vesicles
- Signal transduction processes and cell-to-cell communication and differentiation
- Dendritic spines reformation because of synaptic plasticity and learning