Sponsored by MagritekReviewed by Maria OsipovaJul 7 2025
In materials science, the molecular weight of polymers is a key property that influences various physical characteristics. Previous studies have demonstrated that benchtop NMR can be used to determine polymer molecular weight through two distinct methods:
- end-group analysis, and
- self-diffusion coefficient.
End-group analysis can be conducted via straightforward 1D experiments using 1H, 19F, 29Si, or other nuclei, provided the end-group signal is both observable and well separated from that of the repeating unit.
However, in cases where the signal of the end group cannot be detected either due to overlapping or simply because the signal is too small—such as in polymers with large molecular weight—self-diffusion coefficients are recommended for molecular weight determination.
While each technique can be applied in different scenarios, this article shows that both methods yield precise and comparable results for the same polymer.
The molecular weight of polyphenylsulfone (PPSU) polymer was initially determined using end-group analysis via a 1D 1H spectrum measured on a Spinsolve 90 ULTRA (Fig. 1).
The methoxy end-group signal appears as a singlet at 3.85 ppm, while the signals corresponding to the repeating units resonate within the 7 to 8 ppm range. The degree of polymerization (DP) is calculated using Eq. 1, and the average molecular weight of this PPSU sample is determined to be 8200 g/mol using Eq. 2.
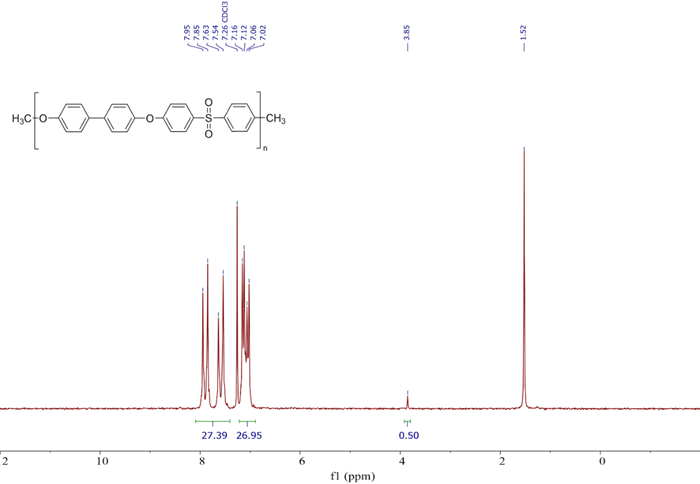
Figure 1. 1D 1H spectrum of PPSU (5 mg/mL in CDCl3) collected on a Spinsolve 90 Carbon ULTRA (32 scans, total measurement time of 4 minutes). Image Credit: Magritek
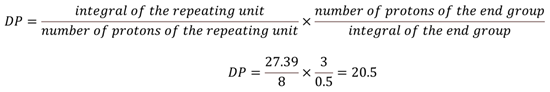 |
Equation 1 |
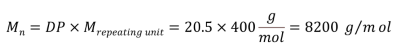 |
Equation 2 |
Polymer molecular weight can also be calculated using self-diffusion coefficients measured through pulsed field gradient NMR, as outlined in a previous article. The diffusion coefficient (D) of the same PPSU sample was measured on the Spinsolve 90 ULTRA, equipped with a pulsed field gradient (0.5 T/m), yielding an average D value of 7.13 x 10-11 m2/second (Fig. 2).
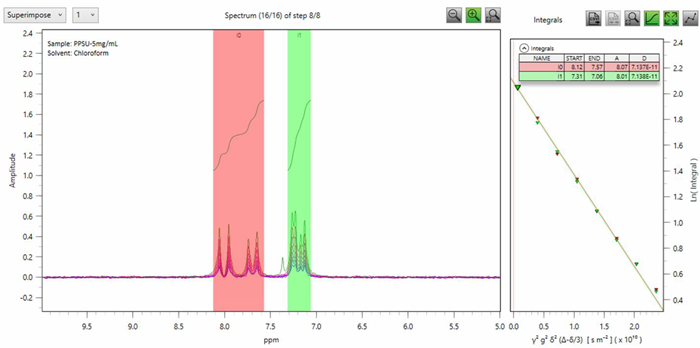
Figure 2. Left: Superimposed plot of the 8 steps PGSTE experiment for PPSU (5 mg/mL in CDCl3). Right: Stejskal-Tanner plot, depicting the signal integral as a function of γ2g2δ2(Δ-δ/3). The data was fitted with a linear function to obtain the diffusion coefficient (D). The total time required to collect this experiment was 15 minutes. Image Credit: Magritek
|
log(D) = log(b’) - νlog(M)
|
Equation 3 |
Using Eq. 3, along with calibration values of -8.23 for log(b’) and 0.49 for v from the universal calibration curve in the publication, an average molecular weight of 8167 g/mol was calculated for PPSU. This value aligns well with the result obtained through end-group analysis.1
Since end-group analysis is an absolute method, the similarity in molecular weight obtained through the diffusion method validates the reliability of the universal calibration curve. This built-in calibration, established using different polymer sets and NMR instruments, ensures consistency across measurements.1
While each molecular weight determination method has distinct benefits and drawbacks depending on the sample, the strong correlation observed in this PPSU study demonstrates that both techniques can be implemented in the Spinsolve benchtop NMR spectrometer.
This allows for molecular weight determination in just minutes, with minimal sample preparation and reduced consumable usage compared to the conventional size exclusion chromatography (SEC) method.
References and Further Reading:
- Voorter, P., et al. (2021). Solvent‐Independent Molecular Weight Determination of Polymers Based on a Truly Universal Calibration. Angewandte Chemie, 134(5). https://doi.org/10.1002/ange.202114536.
This information has been sourced, reviewed and adapted from materials provided by Magritek.
For more information on this source, please visit Magritek.