The actual value of microscopic images is in the data that they provide. One of the major challenges in microscopy is image segmentation as it serves as a foundation for all other image analysis processes. Python-based machine learning methods, such as deep learning and pixel classification, are used in Zen Intellesis to arrive at reproducible and robust segmentation results.
Zen Intellesis is not time-consuming and allows even novice users to train and automatically segment hundreds of images with minimal bias.
Highlights
Use Deep Learning to Segment Images
Zen Intellesis allows users to train the software on a few images and further suitable machine learning algorithms, such as deep learning, are used to segment various classes of objects in several images.
These time-efficient python-based tools utilize pre-trained networks for segmentation and pixel classification. Irrespective of the origin, complex multi-modal data that are also multidimensional can be easily analyzed. Specific deep learning models can also be imported and used with this software.
Enjoy Smooth Workflow Integration
ZEN Intellesis’s software module makes the machine learning algorithms simple to use. For image segmentation, the user should load the image, define classes, give pixel labels and train the model. For tasks such as Nucleus Segmentation, a model that is already trained can be imported, and then the dataset can be segmented and analyzed.
A customized network can also be trained and imported into ZEN. ZEN core Imaging Software and ZEN Blue possess the complete Image Analysis Framework of Intellesis. The integration supports scripting and makes sure that necessary Metadata is available for consequent processing.
Analyze Multi-Modal Images from Many Sources, in Many Formats
Zen Intellesis allows easy segmentation of multidimensional images from various sources such as:
- Superresolution Microscopy
- Widefield Microscopy
- Label-Free Microscopy
- Fluorescence Microscopy
- Light Sheet Microscopy
- Confocal Microscopy
- X-Ray Microscopy
- Electron Microscopy
With Zen Connect and Zen Intellesis, images from different microscopes can be used to extract even more valuable information. Image features from all methods are used at the same time to segment the structures of a selected image. Bio-format compatible images can be segmented by importing TXM images, OME-TIFF or during the usage of a third-party import function.
Application Example Life Sciences
Scratch Assay
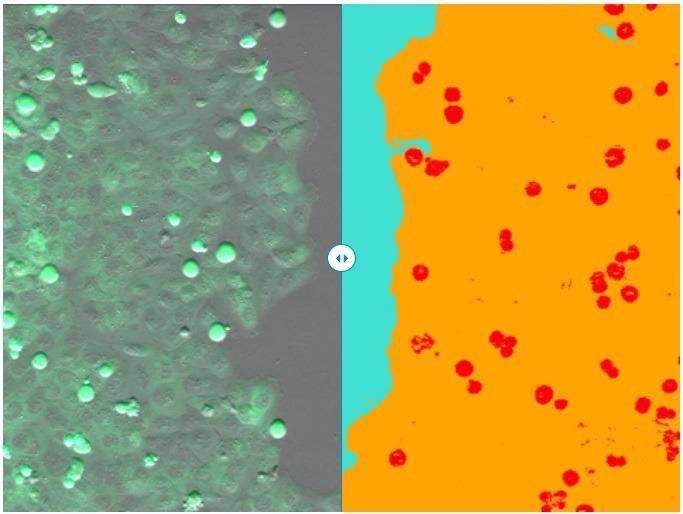
(left) Single frame of recorded time-lapse movie. (right) Segmentation result of ZEN Intellesis - scratch area (turquoise), cell layer (orange), and mitotic cells (red). Image Credit: Carl Zeiss Microscopy GmbH
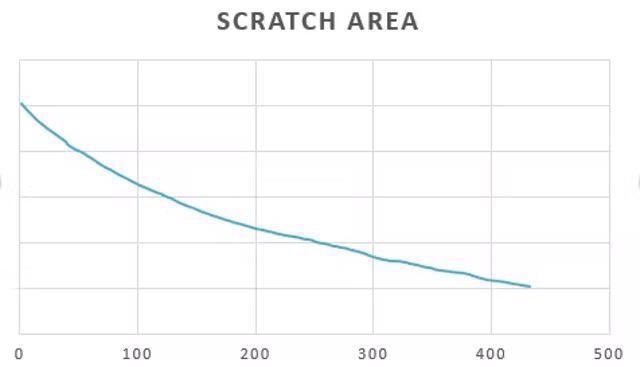
Based on ZEN Intellesis segmentation results, the size of the scratch area was measured over time using the ZEN Image Analysis module. Image Credit: Carl Zeiss Microscopy GmbH
Spines and Dendrites
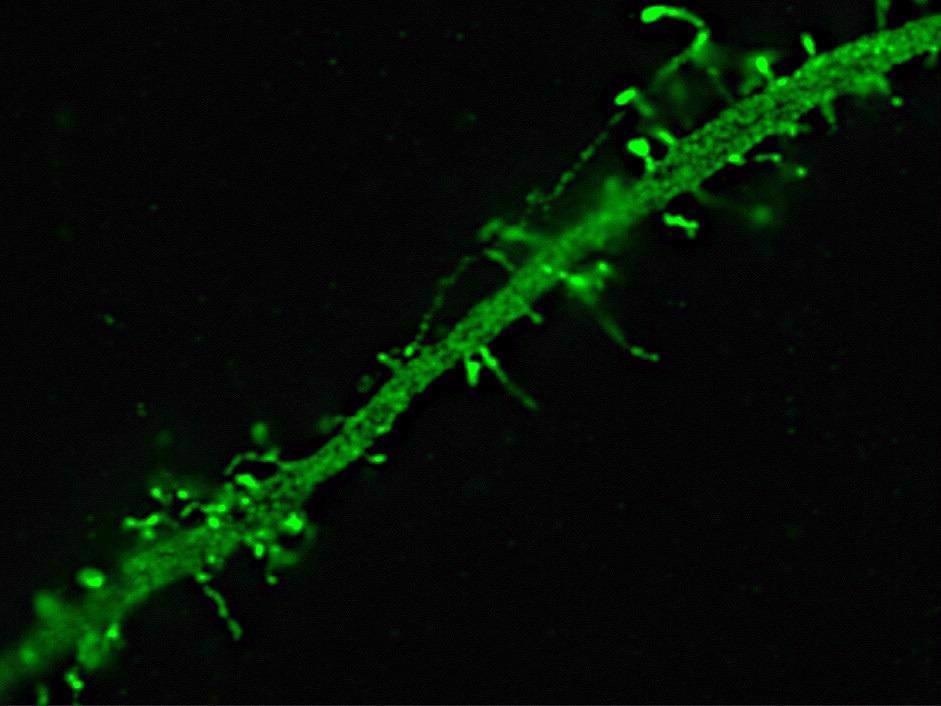
Dendrite of a neuron expressing Green Fluorescent Protein. Image acquired with structured illumination on an Elyra PS.1 showing spines on a dendrite. Image Credit: Carl Zeiss Microscopy GmbH
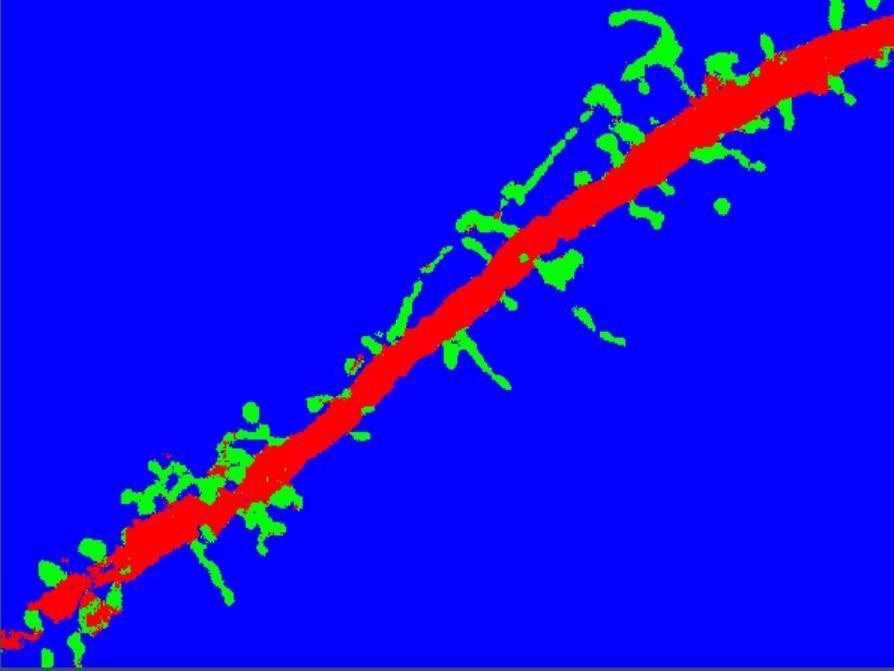
Segmentation with ZEN Intellesis results in a clear separation of spines (green) from dendrite (red) and background (blue). Image Credit: Carl Zeiss Microscopy GmbH
Drosophila
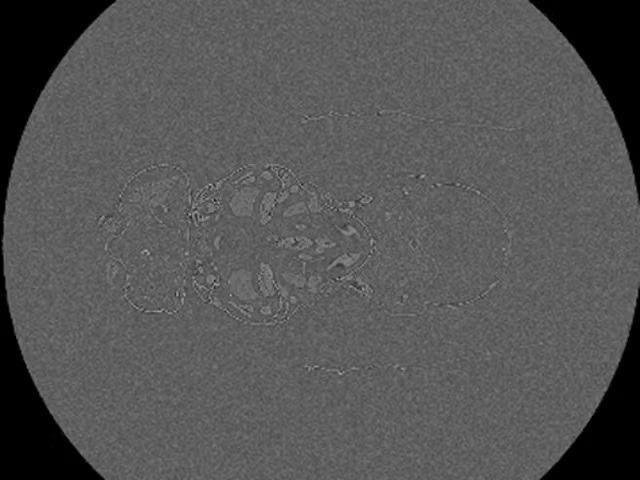
X-Ray micrograph of a Drosophila melanogaster. 1400 slide z-stack of the whole fruit fly acquired with ZEISS 520 Versa. Image Credit: Carl Zeiss Microscopy GmbH
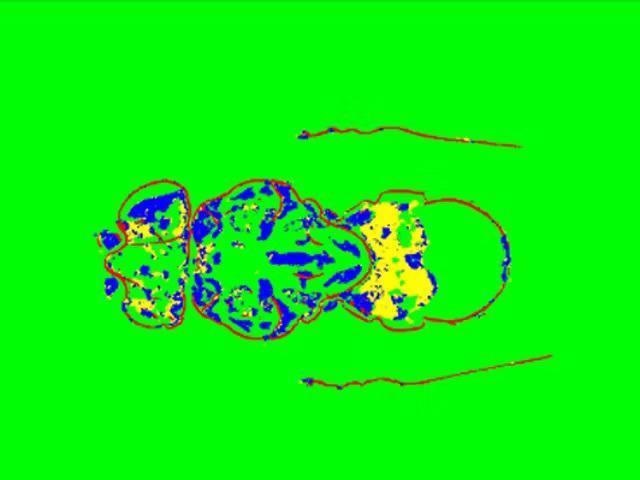
Segmentation result obtained with ZEN Intellesis – exoskeleton (red), inner structures (blue and yellow), background (green). Image Credit: Carl Zeiss Microscopy GmbH

Rendered 3D model based on exoskeleton class. Image Credit: Carl Zeiss Microscopy GmbH
Application Example Materials Analysis
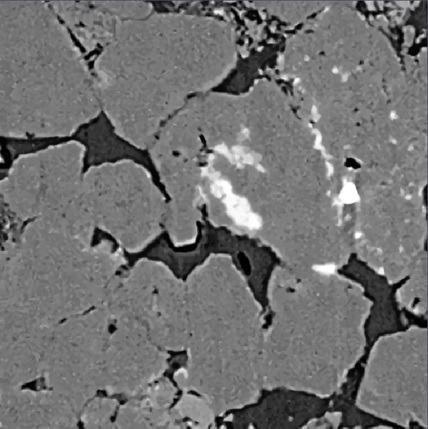
XRM Sandstone – Original. Image Credit: Carl Zeiss Microscopy GmbH
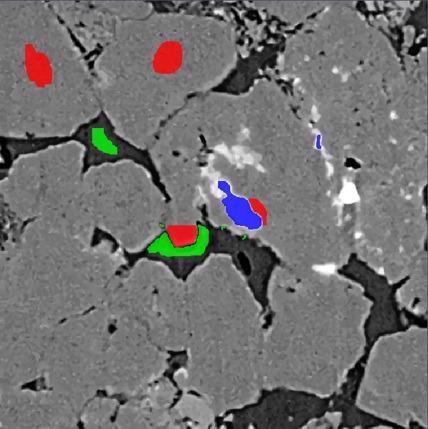
XRM Sandstone – Labeled. Image Credit: Carl Zeiss Microscopy GmbH
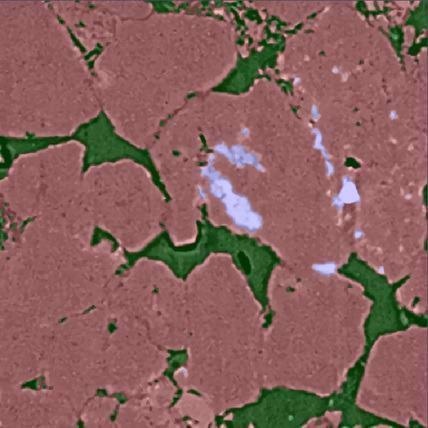
XRM Sandstone – Trained & Segmented. Image Credit: Carl Zeiss Microscopy GmbH

XRM Sandstone – IP Function Output. Image Credit: Carl Zeiss Microscopy GmbH