The bonding interaction amongst Influenza A and the peptide "PeB," which selectively binds the viral surface protein hemagglutinin, has been investigated using electrically controlled deoxyribonuclic acid (DNA) nanolevers in the journal Advanced Materials.
Study: Measuring Influenza A Virus and Peptide Interaction Using Electrically Controllable DNA Nanolevers. Advanced Materials Technologies.Image Credit: Jarun Ontakrai/Shutterstock.com
PeB is conjugated to DNA strands that are bonded to complementary anchors and fixed on the electrode surface of a "switchSENSE" biochip. A fluorophore is attached to the surface-tethered DNA strand, whereas the complementary strand has a multivalent configuration containing up to three PeB peptides. A negative voltage is used to keep the nanolevers erect (static).
As the current epidemic of the SARS-CoV-2 virus has demonstrated, viral pandemics represent a tremendous threat to humanity. A similar pandemic, known as the Spanish flu, was induced by an influenza A virus and led to millions of fatalities around the world. Annual outbreaks of different intensity are still caused by influenza A viruses. The globe was recently confronted with the swine flu in 2009.
Influenza A is an Orthomyxoviridae virus that is encased in a lipid bilayer that contains three cell membranes: hemagglutinin (HA), neuraminidase (NA), and the M2 proton channels. Neuraminidase is a glycoside cleaver that is essential for the discharge of viral particles from infected cells.
Subsequent to viral fusing, hemagglutinin binds to the host cell's sialic acid (SA)-containing cellular receptors. Understanding viral internalization and infection requires kinetic analysis of affinity and fervency coefficients of various receptor binding to hemagglutinin.
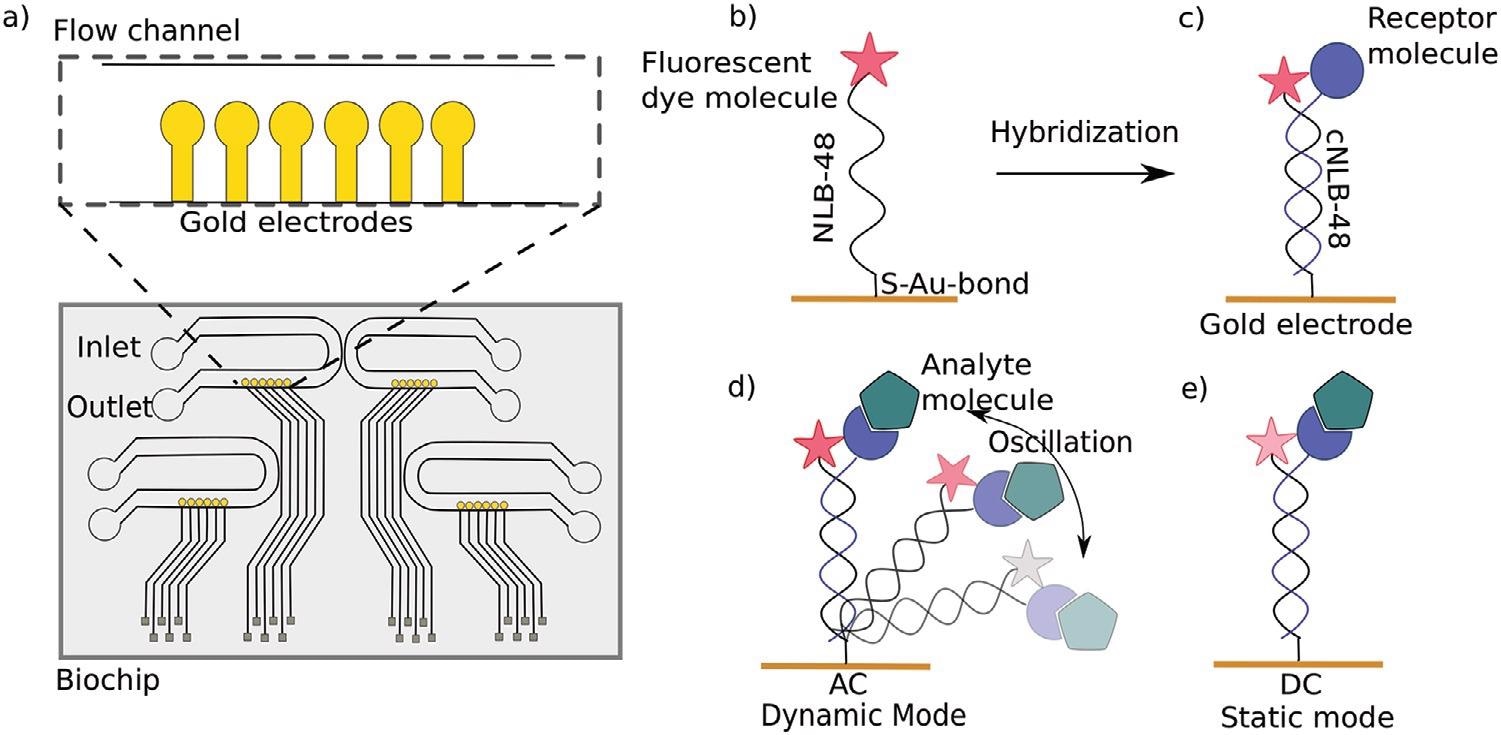
a) Top view of a switchSENSE microfluidic biochip with four flow channels. Each channel comprises six gold electrodes. The more detailed view shows the six independent measurement electrodes in a row within the microfluidic channel. b) Schematic overview of a single stranded DNA nanolever (NL-B48) immobilized on a gold electrode via thiol coupling. The nanolever carries a fluorophore at the lateral end. c) The DNA monolayer is functionalized with the ligand of interest by hybridization of the complementary DNA strand carrying a receptor molecule at the distal end. d) Dynamic measurement mode: The double stranded DNA nanolever is actuated to a switching motion by applying an alternating potential. The fluorescence signal is gradually quenched by energy transfer upon approaching the gold surface. Read-out of this mode is the switching speed of DNA nanolevers. The switching speed is slowed down upon binding of an analyte to the ligand molecule adding friction to the nanolever. e) Static measurement mode: DNA nanolevers are kept at an upright position. Read-out of this mode is the change in fluorescence intensity upon binding of an analyte due to changes in the local environment of the dye. Drawings are not to scale. Image Credit: Kruse M et al., Advanced Material
The ability to calculate not just the dissociation constant but also rate constants is one of the key features of the switchSENSE technique. These kinetic metrics are becoming increasingly important in determining the effectiveness of candidates in medication development.
Other methods for determining dissociation constants exist, but they lack the capacity to resolve bonding kinetic parameters. Radioligand binding tests, microscale thermophoresis (MST), and isothermal calorimetry are some examples of these.
Surface plasmon resonance is one technology that allows for real-time monitoring of kinetic parameters (SPR). SPR measures and switchSENSE technologies are similar in that they both have benefits and drawbacks. As a result, a similar naming system is used.
The quantifiable metrics of kinetic parameters in a timely manner, label-free detection, minimal sample expenditure, and high sensitivity of the technology are all benefits of SPR and switchSENSE. The disadvantages of both procedures are that one of the participants must be immobilized, which may have an impact on binding behavior, and that mass transfer is limited.
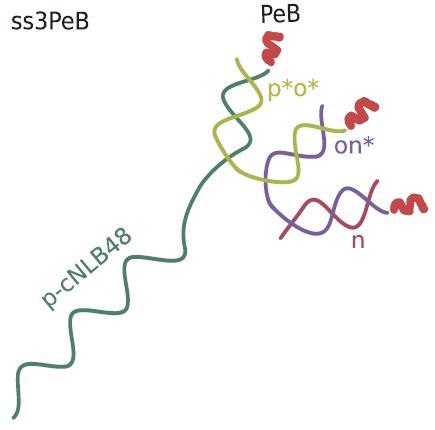
Schematic view of the nanoconstruct ss3PeB (not drawn to scale). The single stranded overhang is complementary to NL-B48 on the electrode surface. Image Credit: Kruse M et al., Advanced Material
The changeable receptor surface is a key benefit of switchSENSE technology over SPR. The switchSENSE sensor surfaces allow ligands to be placed at a specific density, eliminating crowding and enabling two distinct ligands to be distributed at a unique ratio to imitate cooperative effects.
Furthermore, because it uses a reversible DNA hybridization immobilization approach, the general sensor surface may be employed for a variety of interactions. switchSENSE is particularly useful when the receptors of interest are DNA-based, as in our situation with DNA-peptide nanoconstructs.
The resultant double-stranded DNA is negatively charged by nature and may be electrically controlled. When an alternating voltage is provided to the electrode surface, the end-tethered DNA strand oscillates (measuring mode called "dynamic mode"), and when a direct voltage is applied, the DNA nanolever stands erect (quantification mode called "static method").
The fluorophore's emission system provides information about the binding event in both circumstances. After a binding event, the speed of the DNA nanolever's motion is delayed by increasing hydrodynamic friction, causing a delay between the electrical and optical signals in the operating mode.
This implies that after imprinting, a DC voltage is applied, resulting in the DNA-peptide constructions being erect and static. Fluorescent proximity detection is then used to examine virus binding. The corresponding second DNA strand has a four-armed shape for this purpose.
On the gold electrode, one arm is partly complementary to the NL-B48 single-stranded DNA (ssDNA). Each of other three arms can be modified to carry the peptide PeB. As confirmed by microscale thermophoresis, the azide-based copper-free click chemical conjugation preserves the peptide's capacity to bind to hemagglutinin.
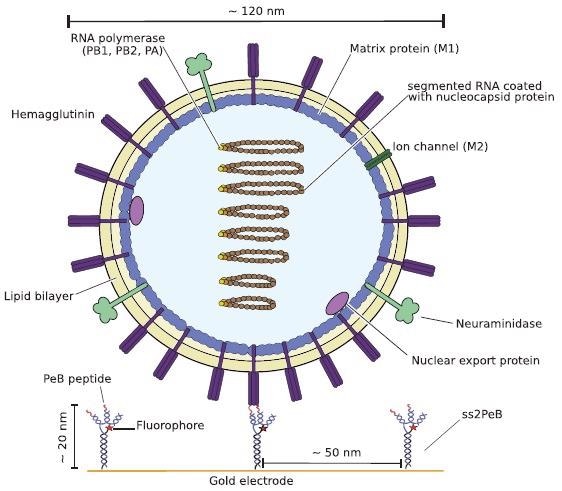
Schematic illustration of one Influenza A virus X31 binding to the peptides attached to DNA nanoconstructs (here shown for ss2PeB). The virus is captured on the sensor surface by specific interaction of hemagglutinin to the presented peptide construct. Schematic is drawn to scale. Image Credit: Kruse M et al., Advanced Material
Because all coupled nanoconstructs must dissolve at the same time for total viral dissociation, this greatly helps stabilize virus capture. Because the virus is still linked to the surface, rebinding will happen if only one pair of participants unbinds, enhancing the local concentration of accessible hemagglutinin proteins (avidity effect).
Due to the combined effect of oligovalent binding and rebinding, this results in a condition where the virus is mostly attached to the nanoconstructs' peptides.
In order to improve repeatability, the impact of chip degradation following tests, which limits the number of accessible nanolevers, must be addressed. Limitations in mass movement can be overcome by lowering the number of receptors and boosting flow rates.
References
Kruse, M., et al. (2021). Measuring Influenza A Virus and Peptide Interaction Using Electrically Controllable DNA Nanolevers. Advanced Materials Technologies. First published: 29 October 2021. https://onlinelibrary.wiley.com/doi/10.1002/admt.202101141
Disclaimer: The views expressed here are those of the author expressed in their private capacity and do not necessarily represent the views of AZoM.com Limited T/A AZoNetwork the owner and operator of this website. This disclaimer forms part of the Terms and conditions of use of this website.