Isothermal titration calorimetry (ITC) can measure heat emission or absorption during binding interactions between proteins and low-molecular weight (LMW) ligands, a focus area of drug discovery and academic research.
With this ability of ITC, protein-small molecule interactions can be studied without the need for immobilization or labeling. Characterizing the variations in enthalpic and entropic contributions to binding of ligands is another key application of ITC.
The New MicroCal PEAQ-ITC System
Broad affinity range is a major challenge involved in the characterization and quantification of binding interactions and therefore needs to be addressed. In a typical drug discovery project, the range of binding affinity values of tiny molecules binding to a target protein varies from low millimolar to subnanomolar. For academic research, an equally broad affinity range is significantly based on the biochemical system being analyzed.
Characterization of binding interactions can be greatly simplified with the combination of appropriately designed analyzes and highly sensitive ITC instruments. The new Malvern Panalytical MicroCal PEAQ-ITC system has improved signal stability, signal-to-noise, and mixing characteristics (Figure 1).
These qualities in conjunction with a sophisticated analysis design feature incorporated into the user-friendly data analysis software optimize analyzes of challenging interactions. This article discusses the advantages of using the MicroCal PEAQ-ITC instrument and software in the investigation of binding interactions by means of direct and competitive titrations.
Improved Data Quality with MicroCal PEAQ-ITC
This experiment compared the new MicroCal PEAQ-ITC against the earlier version, MicroCal iTC200, by performing a titration of acetazolamide with bovine carbonic anhydrase II.
The results reveal that short-term noise in the case of the MicroCal PEAQ-ITC is five times lower than the MicroCal iTC200.
This improved data quality raises confidence while studying challenging data with low heats or when a sample is expensive or low concentrations are needed for high-precision quantitation of low-nanomolar affinity interactions.
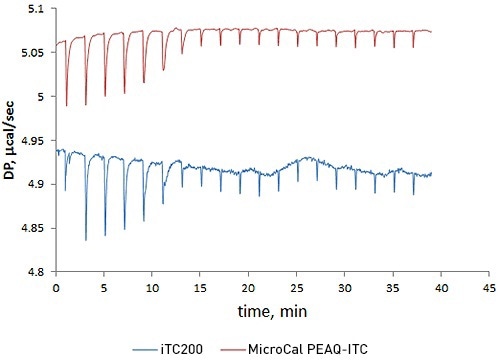
Figure 1. Overlay of raw ITC data for the titration of 20µM acetazolamide (ACZA) into 2µM bovine carbonic anhydrase II (bCAII) using the MicroCal PEAQ-ITC (top) and MicroCal iTC200 (bottom) systems.
Broad Range of Binding Affinities
Titrations of bovine carbonic anhydrase II (bCAII) and human carbonic anhydrase I (hCAI) were performed with a number of inhibitors to demonstrate the suitability of the new PEAQ MicroCal ITC system for characterization of binding interactions. Figure 2 presents the typical data sets and Table 1 summarizes the results.
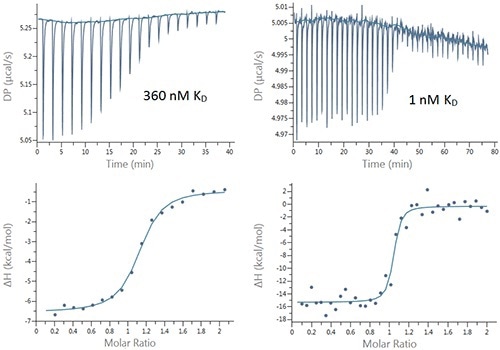
Figure 2. Raw (top) and integrated heat (bottom) plots for the titration of 200µM Furosemide into 20µM bCAII (left) using 2µL injections and 25µM ethoxzolamide into 2.5µM bCAII (right) using 1µL injections.
Table 1. Summary of the results for the titrations (n=3) of hCAl and bCAII with a series of inhibitors.
| Protein |
cmpd, n=3 |
N (sites) |
error (N), % |
KD (M) |
pKD |
error (KD), % |
ΔH (kcal/ mole) |
error (ΔH), % |
| hCAI |
Sulfanilamide |
1.0 |
fixed |
3.2E-04 |
3.5 |
16 |
-5.9 |
15 |
| Ethoxzolamide |
1.0 |
1.0 |
1.9E-08 |
7.7 |
7 |
-8.7 |
1 |
| bCAII |
Furosemide |
1.0 |
1.7 |
3.6E-07 |
6.4 |
18 |
-6.3 |
3 |
| Acetazolamide |
0.9 |
0.6 |
1.8E-08 |
7.7 |
3 |
-12.6 |
3 |
| Ethoxzolamide |
0.9 |
0.3 |
4.4E-10 |
9.4 |
95 |
-14.4 |
3 |
Ethoxzolamide Vinj=1 uL
|
1.0
|
1.0
|
1.3E-09
|
8.8
|
76
|
-15.1
|
1
|
All the interactions were analyzed utilizing a standard procedure of injecting 18.2µL aliquots of the ligand into the protein solution within the cell. The errors in the KD were all below 20% and less than 10% in some cases for the interactions with affinities of 18nM and weaker.
The errors in the enthalpy and stoichiometry data were also all below 3% other than for the sulfanilamide. These values reveal the ability of the PEAQ MicroCal ITC system to produce highly reproducible data even with low protein concentrations down to 2.5µM.
Although KD value varied drastically for the ethoxzolamide, the tightest binder, the error of 94% is fair enough for these tight interactions, providing a certain amount of confidence in the data. The low signal-to-noise and high accuracy of the pipette of the new MicroCal PEAQ-ITC instrument enable using injection volumes as small as 1µL.
This approach enables populating the transition region with more data points, thereby improving the affinity data quality (Figure 2). The data presented in Figure 3 are in terms of pKD, which is possibly the more appropriate method for affinity data comparison.
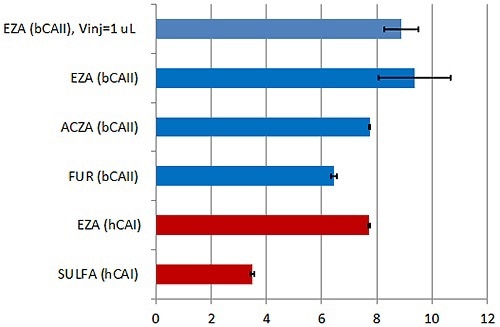
Figure 3. Summary of the KD values obtained with the new MicroCal PEAQ-ITC instrument and analysis software for a series of LMW inhibitors of hCAI (red) and bCAII (blue). Error bars indicate errors in units of pKD (errors in % of KD are given in Table 1).
Competition Titration
A competition experiment involves injecting a strong binding ligand into the target protein under the presence of a weaker, competitive compound. For an ITC titration, an adequate number of data points won’t be present in the transition region when the weaker inhibitor is absent. It is necessary to have an adequate number of data points in the transition region in order to fit a unique binding isotherm.
Figure 4A shows an injection of 200 µM ethoxzolamide into 20 µM bCAII in 2µL aliquots. Ideal experimental conditions that would ensure a high enough heat value for better signal to noise and an adequate number of data points in the transition range can be achieved using the competition experiment design tool equipped with the MicroCal PEAQ-ITC (Figure 5). Figure 4B shows the resulting titration.
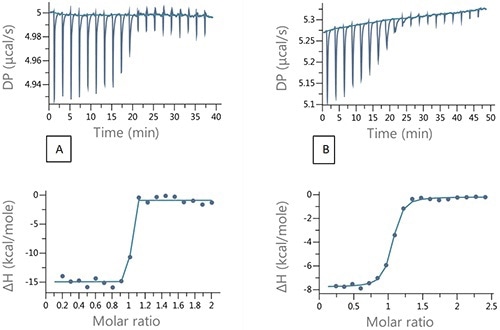
Figure 4. Raw and integrated ITC data for (A) direct titration of 200 µM EZA into 20µM bCAII and (B) Competition titration 200 µM EZA into 20µM bCAII and 100 µM FUR.
The results summarized in Table 2 show good agreement between the binding parameters for the tight binder obtained from the competition titration and the data from direct titration, demonstrating the applicability of the competition experiments and the design tool for characterization of tight binding interactions.
Table 2. Binding parameters of bCA II interaction with EZA obtained in direct titration of 25µM EZA into 2.5µM bCA II and in a competition titration of 200µM EZA into 20µM bCA II premixed with 100µM FUR, Vinj=2µL.
| Experimental setup |
KD (nM) |
error(KD), % |
ΔH (kcal/mol) |
error(DH), % |
| Direct titration |
0.4 |
95 |
-14.4 |
3 |
| Competition titration |
0.4 |
40 |
-13.9 |
6 |
Conclusion
The results demonstrate the ability of the new Microcal PEAQ-ITC system to provide data with improved quality. The instrument can quantify a wide range of affinities due to its high signal-to-noise ratio.
The results also demonstrate the outstanding repeatability of the system for measurement of nanomolar affinities. In addition, the error of roughly 20% or better for micromolar interactions showed the unprecedented repeatability for calculating KD.
The use of competition experiments extends the affinity range of ITC into the high picomolar range. The intricacy of the experimental design is a major shortcoming of that approach. The simulation tool in the MicroCal PEAQ-ITC Analysis Software simplifies the design and analysis.
Besides improved signal-to-noise ratio, completely automated data analysis, reduced user subjectivity in corroborating data quality, and shortened analysis time are the advantages of the MicroCal PEAQ-ITC system.
The ability to determine data quality and perform fitting within few seconds per analysis enables the instrument to analyze large data sets of 50 or more analyzes within a short period of time.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.