The physical properties and behavior of polymers and polysaccharides are strongly based on the properties of their molecules. Molecular weight, molecular weight distribution, molecular structure and size all impact the behavior of the material. Gel permeation chromatography (GPC), also known as size-exclusion chromatography (SEC), is the most commonly used tool for assessing these parameters.
The GPC principle involves separation of the sample as it travels through an inert but porous column matrix. Larger molecules are excluded while smaller molecules penetrate the pores more deeply. The result is a separation based on hydrodynamic volume, but the desire is to know the molecular weight of the sample.
The molecular weight was determined by comparing the elution time of the sample to that of standards of known molecular weight. Presently however, a light scattering detector is a common tool that permits polymer molecular weight to be determined independent of retention time.
Measuring Rg using multi-angle light scattering and intrinsic velocity simultaneously offers excellent insight into the structure of natural and synthetic polymer molecules as well as their molecular weight.
The Viscotek SEC-MALS 20
The Viscotek SEC-MALS 20 as shown in Figure 1 is a 20° angle light scattering device that can measure Rg and molecular weight. It can be used as part of a multi-detector GPC system that integrates light scattering with other detectors such as refractive index (RI), ultraviolet (UV) and intrinsic viscosity (IV) to generate a large amount of information about a sample simultaneously.
In this article, molecular weight data from the Viscotek SEC-MALS 20 is combined with measurements of Rg and IV to study the structure of different polysaccharides including pullulan, dextran, hydroxypropyl cellulose (HPC), pectin and gum Arabic. The differences between conformation and Mark-Houwink plots are also studied.

Figure 1. The Viscotek SEC-MALS 20 system SEC-MALS 20 application note - MRK1929-01 2.
Materials and Methods
The SEC-MALS 20 detector was connected to a Viscotek GPCmax and a Viscotek TDA system. Samples were separated using 2 x Viscotek A6000M columns (Dextran, Pectin, Gum Arabic) or one column (Pullulan, Hydroxypropyl cellulose). The mobile phase was phosphate buffered saline. The SEC-MALS 20 system was calibrated with a 19kDa PEO standard. The columns and detectors were all held at 35°C to ensure a good separation and maximize baseline stability.
All samples were dissolved in the mobile phase to a concentration of between 0.5 and 3 mg/ml and a 100 µl injection volume was used. The Dextran sample has a known molecular weight and was used to check the calibration. All samples were allowed to dissolve overnight to ensure full dissolution.
Results
All of the samples were successfully separated on the system. Overlaid RI chromatograms are shown for the pectin, gum Arabic and dextran (figure 2A) and for the pullulan and HPC (figure 2B).
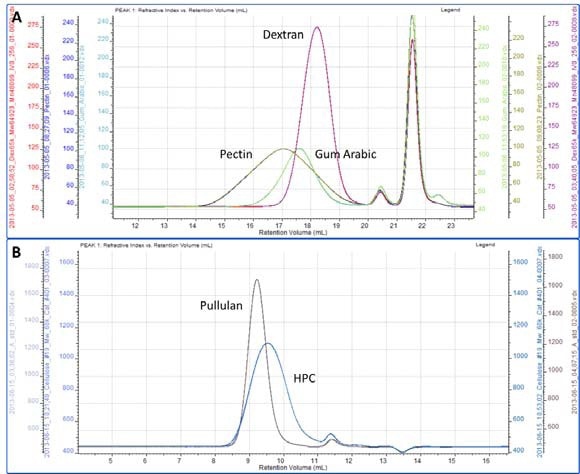
Figure 2. A. Overlaid RI chromatograms are pectin, dextran and gum Arabic. B. Overlaid RI chromatograms or pullulan and HPC.
As a more comprehensive example, a chromatogram of pectin is shown in figure 3A showing I, LS (90°) and IV detectors. Figure 3B shows the angular data from the SEC-MALS 20 detector. A slight angular dependence can be observed across the different angles allowing Rg to be measured for this sample. Figure 3C shows the RI peak overlaid with the measured molecular weight, Rg, IV and Rh.
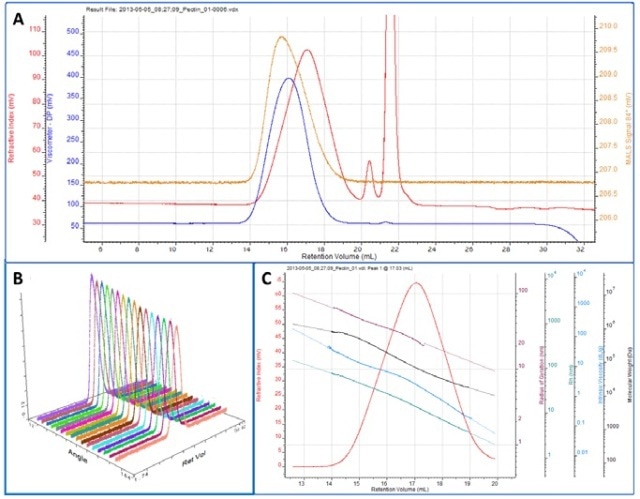
Figure 3. A. Chromatogram showing RI (red), LS (90°) (orange) and IV (blue) detector responses for pectin. B. Plot showing the MALS data. C. Derived data showing RI peak overlaid with Rg (maroon), molecular weight (black), IV (turquoise), Rh (teal).
Table 1. Measured molecular weight distributions, IV, Rh and Rg values for the different polysaccharide samples.
| Sample Id |
Dextran |
Pectin |
Gum Arabic |
Pullulan |
HPC |
| Injection |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
1 |
2 |
| Mn - (kDa) |
51.42 |
51.56 |
51.07 |
52.40 |
292.81 |
291.42 |
185.77 |
187.28 |
61.86 |
58.06 |
| Mw - (kDa) |
63.18 |
63.34 |
115.07 |
115.89 |
515.13 |
519.06 |
196.57 |
197.49 |
83.03 |
81.86 |
| Mz - (kDa) |
77.84 |
78.29 |
249.56 |
252.81 |
974.47 |
1,000.00 |
210.70 |
210.47 |
118.79 |
125.51 |
| IV - (dl/g) |
0.256 |
0.256 |
3.671 |
3.689 |
0.158 |
0.156 |
0.674 |
0.678 |
1.050 |
1.066 |
| Rh - (nm) |
6.06 |
6.07 |
16.97 |
17.06 |
10.26 |
10.21 |
16.63 |
16.70 |
13.98 |
13.93 |
| Rg - (nm) |
7.55 |
7.09 |
25.83 |
25.98 |
8.08 |
8.55 |
16.42 |
16.58 |
22.48 |
20.03 |
Table 1 summarizes the results for the different samples. Duplicate injections were done and it can be seen that the repeatability between injections is excellent across all of the measurements. While the study of the numerical results is an excellent quantitative way to compare samples, plotting the data on Mark-Houwink and Conformation plots is an ideal way to compare samples quickly from a visual perspective.
A Mark-Houwink plot shows intrinsic viscosity as a function of molecular weight whereas a conformation plot shows Rg as a function of molecular weight. Both of these are therefore a way of visualizing the increase in size as molecular weight increases. They allow immediate comparison of different samples with respect to each other and are ideally suited to the study of branching and other structural changes. In this case though, we are just interested in seeing where the different polysaccharides fall on the plots.
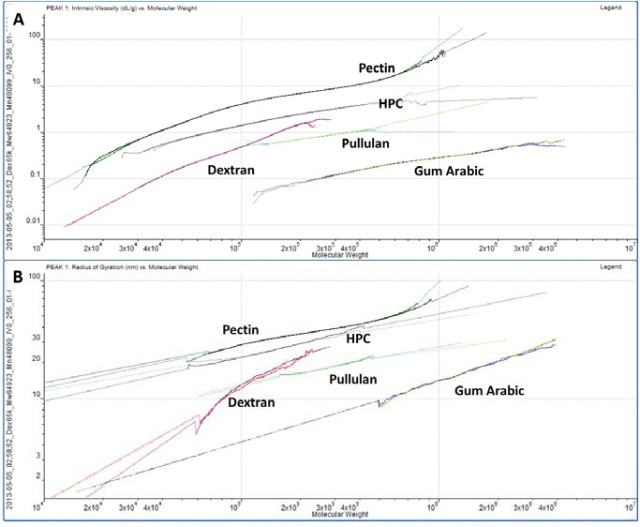
Figure 4. A. Overlaid Mark-Houwink plots for the polysaccharides. B. Overlaid conformation plots for the polysaccharides. Solid lines indicate real data and dotted lines indicate extrapolated data.
Discussion
The information contained within the results becomes very clear while studying the Mark-Houwink and the Conformation plots. While gum Arabic is the highest molecular weight sample, it is also the ‘lowest’ on both plots indicating that for its mass, it is the smallest (conformation plot) or most dense (Mark-Houwink plot) of the molecules under study. Although gum Arabic is primarily a polysaccharide, it is associated with a protein component which adds mass with just a small increase in size as this is as expected. On the other hand, pectin has the longest line in both plots indicating that this is the most polydisperse molecule of the ones studied.
This is compliant with the fact that it has the widest peak in the chromatogram. This is clearer on the Mark- Houwink plot than on the conformation plot. The conformation plot is restricted to molecules over approximately 10nm radius where the light scattered by the samples is anisotropic. The pectin distribution includes certain material below the threshold Rg cannot be measured. IV is not restricted by size so it can measure the entire molecular weight distribution. As the ‘highest’ molecule on both plots, pectin identifies itself as the largest conformation plot and least dense Mark-Houwink plot molecule of those under investigation.
HPC, pullulan and dextran all fall in between the pectin and gum Arabic samples showing that their structures are somewhere between these two extremes. While the two plots offer similar information, it is worth understanding that each has weaknesses and strengths that will make their appropriateness application dependent. The conformation plot as discussed above cannot be used for smaller molecules. However as a direct size measure it does not depend on any shape models. If IV is to be converted into a size value, some shape model must be assumed, as in the calculation of Rh in Table 1.
Conclusion
In summary, in this application note, the molecular weight, size and intrinsic viscosity of a selection of polysaccharides were determined using the Viscotek TDA and SEC-MALS 20 systems. Conformation and Mark-Houwink plots were then drawn which both clearly demonstrate the different structures between the different samples. The differences in molecular structures between the different polysaccharides are very clear and can be easily observed from the data. With single detector GPC systems, this characterization level was not possible previously with single detector GPC systems.
The use of light scattering and intrinsic viscosity detectors in conjunction with GPC allows detailed characterization of natural and synthetic polymer samples.

This information has been sourced, reviewed and adapted from materials provided by Malvern Panalytical.
For more information on this source, please visit Malvern Panalytical.