Jul 4 2019
A group of scientists from Brown University, headed by Jacob Rosenstein, encoded and decoded images of an Egyptian cat, an ibex, and an anchor from mixtures of small molecules known as metabolites.
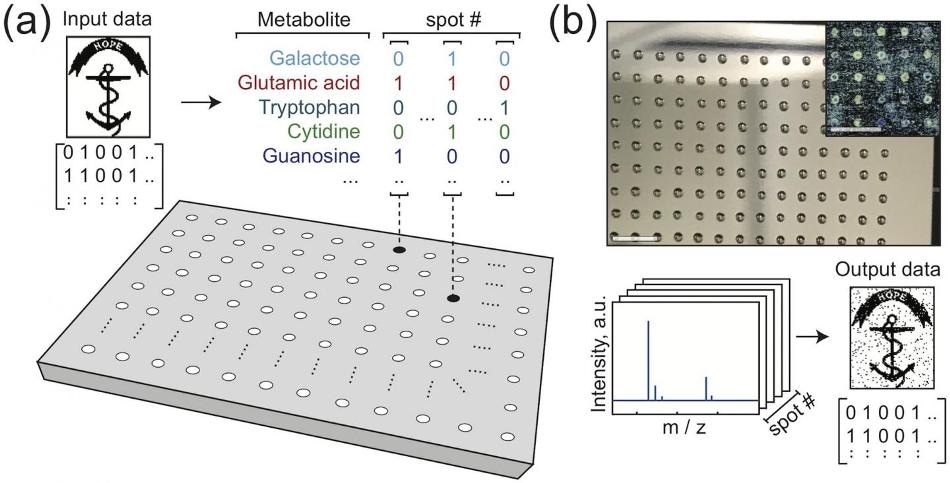 Writing and reading data encoded in mixtures of metabolites. (a) Binary image data is mapped onto a set of metabolite mixtures, with each bit determining the presence/absence of one compound in one mixture. For example, a spot mapped to four bits with values [0 1 0 1] may contain the 2nd and 4th metabolite at that location. (b) Small volumes of the mixtures are spotted onto a steel plate and the solvent is evaporated (scale bars: 5 mm). This chemical dataset is analyzed by MALDI mass spectrometry (b, bottom). Using the observed mass spectrum peaks, decisions are made about which metabolites are present. These decisions are assembled from the array of spots to recover the original image. The image shown is the Rhode Island Hope Regiment Colors [28]. (Image credit: Kennedy et al., 2019)
Writing and reading data encoded in mixtures of metabolites. (a) Binary image data is mapped onto a set of metabolite mixtures, with each bit determining the presence/absence of one compound in one mixture. For example, a spot mapped to four bits with values [0 1 0 1] may contain the 2nd and 4th metabolite at that location. (b) Small volumes of the mixtures are spotted onto a steel plate and the solvent is evaporated (scale bars: 5 mm). This chemical dataset is analyzed by MALDI mass spectrometry (b, bottom). Using the observed mass spectrum peaks, decisions are made about which metabolites are present. These decisions are assembled from the array of spots to recover the original image. The image shown is the Rhode Island Hope Regiment Colors [28]. (Image credit: Kennedy et al., 2019)
The capability of this small-molecule information storage system has been described in a new paper published on July 3rd, 2019, in the open-access journal PLOS ONE.
Microchips have turned out to be the typical mode for data storage, for instance, a snapshot on a phone or a document on a home computer. Recently, researchers worked on methods to encode information using biomolecules, like DNA, by producing an artificial genome. In this new research, the scientists demonstrate that information can be encoded using another kind of biomolecule called metabolites.
They employed liquid-handling robots to record digital information by dotting mixtures of metabolites into a grid on a surface. The sites and identities of the metabolites, interpreted by an instrument known as a mass spectrometer, reported out binary data. This technique enabled them to encode the information from an image and subsequently decode it to redraw the image with an accuracy of 98%–99.5%.
Mixtures of metabolites, known as metabolomes, provide several benefits over genomes for recording information. Metabolites are more varied, smaller, and have the capacity to store information at larger density.
This proof-of-principle study demonstrates that small-molecule information storage can effectively encode over 100,000 bits of digital images into synthetic metabolomes. According to the researchers, with further advancement, the amount and density of information that can be encoded will increase considerably.
We encoded several small digital images into mixtures of metabolites, and read back the data by chemically analyzing the mixtures. A molecular hard drive or a chemical computer might still seem like science fiction, but biology shows us it is possible. We wanted to show in a mathematically precise way how to write and read digital data using some of the small molecules that our bodies use every day.
Jacob Rosenstein, Lead Researcher, Brown University
This study was supported by financial support from the Defense Advanced Research Projects Agency (DARPA W911NF-18-2-0031) to BMR and JKR.