Jul 3 2019
Scientists at Chalmers University of Technology, Sweden, have designed a unique method for analyzing proteins which could pave the way for medicinal research.
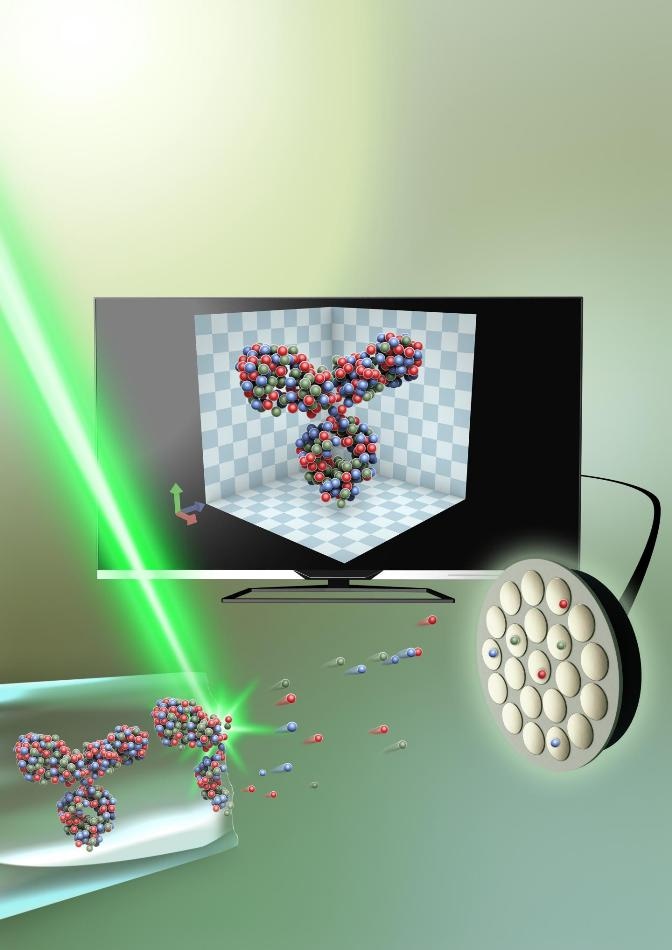 The protein, captured in an extremely thin piece of glass—around 50 nanometers in diameter, is sliced up, atom by atom, with the help of an electrical field. It is then analyzed through atom probe tomography, and the 3D structure is recreated on a computer. (Image credit: Small: Volume 15, Issue 24, Atom Probe Tomography for 3D Structural and Chemical Analysis of Individual Proteins Gustav Sundell, Mats Hulander, Astrid Pihl, Martin Andersson Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.)
The protein, captured in an extremely thin piece of glass—around 50 nanometers in diameter, is sliced up, atom by atom, with the help of an electrical field. It is then analyzed through atom probe tomography, and the 3D structure is recreated on a computer. (Image credit: Small: Volume 15, Issue 24, Atom Probe Tomography for 3D Structural and Chemical Analysis of Individual Proteins Gustav Sundell, Mats Hulander, Astrid Pihl, Martin Andersson Copyright Wiley-VCH Verlag GmbH & Co. KGaA. Reproduced with permission.)
The scientists collected proteins in a nanocapsule made of glass and successfully developed a unique model of proteins in natural environments. The findings have been reported in the scientific journal, Small.
Proteins are target-seeking and perform various tasks that are essential for the survival and functions of the cells. These have gained considerable attention for the development of new medicines—especially those proteins present in the cellular membrane and determine which molecules are allowed to enter the cell and which are not. This shows that understanding the performance of such proteins is a crucial challenge to develop more modern medicines. This, however, is very difficult because such proteins are more complex. At present, although many different methods are used for imaging proteins, none offers a complete solution to the difficulty of studying individual membrane proteins in their natural environment.
A team of researchers at Chalmers University of Technology, led by Martin Andersson at the Department of Chemistry and Chemical Engineering, has successfully used atom probe tomography to image and analyze proteins. Atom probe tomography has existed for some time, but has not formerly been utilized in this way—but instead for studying various other hard materials, including metals.
It was in connection with a study of contact surfaces between the skeleton and implants when we discovered we could distinguish organic materials in the bone with this technique. That gave us the idea to develop the method further for proteins.
Martin Andersson, Department of Chemistry and Chemical Engineering, Chalmers University of Technology
But it is difficult to develop a method to keep the proteins intact in their natural environment. The scientists, however, achieved this by encapsulating the protein in a very thin piece of glass, only about 50 nm in diameter (a nanometer is one/millionth of a millimeter). Later, they removed the glass’ outermost layer by applying electric field, freeing the protein atom by atom. The protein could then be rebuilt in 3D on a computer.
The study outcomes have been confirmed by comparing with the available 3D models of known proteins. The scientists are expected to further develop the method to improve the speed and precision in the future.
This technique is groundbreaking in numerous ways. In addition to modeling the 3D structure, it simultaneously shows the chemical composition of the proteins.
Our method offers a lot of good solutions and can be a strong complement to existing methods. It will be possible to study how proteins are built at an atomic level.
Martin Andersson, Department of Chemistry and Chemical Engineering, Chalmers University of Technology
This method could help in studying all proteins—a feat that is currently not possible. At present, only about 1% of membrane proteins have been structurally analyzed effectively.
“With this method, we can study individual proteins, as opposed to current methods which study a large number of proteins and then create an average value,” said Gustav Sundell, a researcher in Martin Andersson’s research team.
The details of an atom's mass can also be obtained using atom probe tomography.
“Because we collect information on atoms’ masses in our method, it means we can measure the weight. We can then, for example, create tests where medicinal molecules are combined with different isotopes—giving them different masses—which makes them distinguishable in a study. It should contribute to speeding up processes for constructing and testing new medicines,” stated Mats Hulander, a researcher in Martin Andersson’s team.