Nov 17 2009
The goal of an integrated, miniaturized laboratory analysis system, also known as a "lab-on-a-chip," is simple: sample in, answer out. However, researchers wanting to use these microfluidic devices to analyze complex solutions containing particulates or other contaminating materials often find that the first part of the process isn't so easy.
Effective sample preparation from these solutions can be laborious, expensive and time-consuming, involving complicated laboratory methods that must be performed by skilled technicians. This can significantly diminish the benefits associated with using miniaturized analytical techniques. Recent work at the National Institute of Standards and Technology (NIST) could help change that.
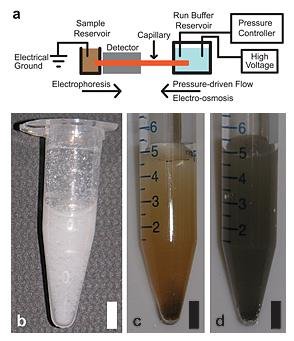 The NIST GEMBE microfluidic sample analysis system is shown schematically in (a) at top. Photographs at bottom show three complex samples that can be successfully analyzed by the technique: (b) whole milk, (c) dirt and (d) coal fly ash. Credit: E. Strychalski, D. Ross, NIST, and A. Henry, ARA Inc.
The NIST GEMBE microfluidic sample analysis system is shown schematically in (a) at top. Photographs at bottom show three complex samples that can be successfully analyzed by the technique: (b) whole milk, (c) dirt and (d) coal fly ash. Credit: E. Strychalski, D. Ross, NIST, and A. Henry, ARA Inc.
NIST researchers Elizabeth Strychalski and David Ross, in collaboration with Alyssa Henry of Applied Research Associates Inc. (Alexandria, Va.), have developed a novel and simple way to analyze samples that are complex mixtures, such as whole milk, blood serum and dirt in solution. In a paper published recently in Analytical Chemistry,* the team describes its latest enhancement to a NIST-developed separation technique called gradient elution moving boundary electrophoresis (GEMBE) (see "New Miniaturized Device for Lab-on-a-Chip Separations" in NIST Tech Beat, Jan. 19, 2007).
GEMBE relies on a combination of electrophoresis and variable pressure-driven flow through a microchannel. Electrophoresis uses electricity to push a mixture in solution through a channel, forcing the individual components to separate as they move at specific rates based on their individual properties, such as size and electrical charge. Complex samples can be difficult to separate cleanly because components in these samples (for example, the fat globules in milk or proteins in blood) can "foul" microfluidic channels in a way that prevents reliable detection of the desired sample components.
The new technique solves this problem by pumping a buffer solution under controlled pressure in the opposite direction. This opposing pressure flow acts as a "fluid gate" between the sample reservoir and the microchannel. Gradually reducing the pressure of the counterflow opens the "gate" a little bit at a time. A specific sample component is detected when the pressure flow becomes weak enough-when the "gate" opens wide enough-that the component's electrophoretic motion pushes it against the pressure flow and into the channel for detection. In this way, different components enter the channel at different times based on their particular electrophoretic motion. Most importantly, the channel doesn't become fouled because the unwanted material in the sample is held out during the analysis by the pressure flow.
In their paper, the researchers validated their GEMBE analysis technique by testing it with solutions of whole milk, dirt, estuarine sediment, coal fly ash, pulverized leaves and blood serum. In all cases-and without the muss and fuss of pre-analysis sample preparation-the system was able to reproducibly separate and quantify specific components from the solutions, including potassium, calcium, sodium, magnesium, lithium and melamine.
"GEMBE is well-suited to the microfluidic analysis of 'real-world' samples," Strychalski says. "We have shown that the method can handle solutions containing particulates, proteins and other materials that would confound the majority of other microfluidic techniques."
Because of its ability to easily and rapidly characterize complex mixtures with minimal preparation, the researchers believe that GEMBE shows enormous promise for diverse applications, such as monitoring contaminants in food or water supplies, determining nutrient levels in soil, detecting biochemical warfare agents, and diagnosing medical conditions. The next steps, they say, are to miniaturize the desktop equipment now used in the system and integrate all of the parts to develop a true "lab-on-a-chip" field analyzer that can rival the effectiveness of a full-scale facility.
* E. Strychalski, A. Henry and D. Ross. Microfluidic analysis of complex samples with minimal sample preparation using Gradient Elution Moving Boundary Electrophoresis. Analytical Chemistry, Vol. 81, No. 24, Dec. 15, 2009; published online Nov. 10, 2009.